MA0597.1_THAP1/Jaspar
| Match Rank: | 1 |
| Score: | 0.62
| | Offset: | 9
| | Orientation: | forward strand |
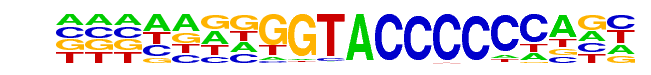
| Alignment: | CCGGTASWGYDGCCYGCABW
---------CTGCCCGCA-- |
|


|
|
PB0024.1_Gcm1_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.54
| | Offset: | 7
| | Orientation: | forward strand |
| Alignment: | CCGGTASWGYDGCCYGCABW---
-------TCGTACCCGCATCATT |
|


|
|
MA0146.2_Zfx/Jaspar
| Match Rank: | 3 |
| Score: | 0.46
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CCGGTASWGYDGCCYGCABW
--GGGGCCGAGGCCTG---- |
|


|
|
MA0469.1_E2F3/Jaspar
| Match Rank: | 4 |
| Score: | 0.45
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CCGGTASWGYDGCCYGCABW
----NNGTGNGGGCGGGAG- |
|


|
|
PB0133.1_Hic1_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.45
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | CCGGTASWGYDGCCYGCABW-
-----GGGTGTGCCCAAAAGG |
|


|
|
PB0052.1_Plagl1_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.44
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CCGGTASWGYDGCCYGCABW
---NNNGGGGCGCCCCCNN- |
|


|
|
PB0117.1_Eomes_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.44
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CCGGTASWGYDGCCYGCABW
GCGGAGGTGTCGCCTC---- |
|


|
|
PB0201.1_Zfp281_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.44
| | Offset: | 6
| | Orientation: | forward strand |
| Alignment: | CCGGTASWGYDGCCYGCABW---
------AGGAGACCCCCAATTTG |
|


|
|
Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 9 |
| Score: | 0.43
| | Offset: | 9
| | Orientation: | forward strand |
| Alignment: | CCGGTASWGYDGCCYGCABW
---------CTTCCNGGAA- |
|


|
|
PB0156.1_Plagl1_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.43
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CCGGTASWGYDGCCYGCABW
---NNNNGGTACCCCCCANN |
|

|
|





















