| p-value: | 1e-16 |
| log p-value: | -3.873e+01 |
| Information Content per bp: | 1.372 |
| Number of Target Sequences with motif | 21.0 |
| Percentage of Target Sequences with motif | 34.43% |
| Number of Background Sequences with motif | 2.6 |
| Percentage of Background Sequences with motif | 3.63% |
| Average Position of motif in Targets | 25.7 +/- 8.2bp |
| Average Position of motif in Background | 21.5 +/- 7.6bp |
| Strand Bias (log2 ratio + to - strand density) | 0.3 |
| Multiplicity (# of sites on avg that occur together) | 1.05 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
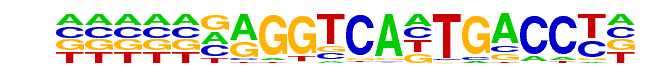
FXR(NR/IR1)/Liver-FXR-ChIP-Seq(Chong et al.)/Homer
| Match Rank: | 1 |
| Score: | 0.46
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | TCHVNSAGBYYVVNGAMBKS
-----NAGGTCANTGACCT- |
|

|
|
POL009.1_DCE_S_II/Jaspar
| Match Rank: | 2 |
| Score: | 0.45
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TCHVNSAGBYYVVNGAMBKS
---CACAGN----------- |
|


|
|
Mouse_Recombination_Hotspot/Testis-DMC1-ChIP-Seq(GSE24438)/Homer
| Match Rank: | 3 |
| Score: | 0.44
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | TCHVNSAGBYYVVNGAMBKS----
----GAAGTANCACGAATNMRAGT |
|


|
|
POL010.1_DCE_S_III/Jaspar
| Match Rank: | 4 |
| Score: | 0.43
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | TCHVNSAGBYYVVNGAMBKS
-----CAGCC---------- |
|


|
|
VDR(NR/DR3)/GM10855-VDR+vitD-ChIP-Seq(GSE22484)/Homer
| Match Rank: | 5 |
| Score: | 0.42
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | TCHVNSAGBYYVVNGAMBKS----
----NGAGGTCANNGAGTTCANNN |
|


|
|
PB0004.1_Atf1_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.41
| | Offset: | 9
| | Orientation: | reverse strand |
| Alignment: | TCHVNSAGBYYVVNGAMBKS-----
---------NCGATGACGTCATCGN |
|


|
|
MA0074.1_RXRA::VDR/Jaspar
| Match Rank: | 7 |
| Score: | 0.41
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | TCHVNSAGBYYVVNGAMBKS
----TGAACCCGATGACCC- |
|


|
|
CRE(bZIP)/Promoter/Homer
| Match Rank: | 8 |
| Score: | 0.40
| | Offset: | 10
| | Orientation: | forward strand |
| Alignment: | TCHVNSAGBYYVVNGAMBKS--
----------CGGTGACGTCAC |
|


|
|
JunD(bZIP)/K562-JunD-ChIP-Seq/Homer
| Match Rank: | 9 |
| Score: | 0.39
| | Offset: | 10
| | Orientation: | reverse strand |
| Alignment: | TCHVNSAGBYYVVNGAMBKS--
----------NGATGACGTCAT |
|


|
|
Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 10 |
| Score: | 0.39
| | Offset: | 7
| | Orientation: | forward strand |
| Alignment: | TCHVNSAGBYYVVNGAMBKS
-------SCCTAGCAACAG- |
|


|
|





















