NPAS2(HLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer
| Match Rank: | 1 |
| Score: | 0.66
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATCAAWTGGA
GTCACGTGGM |
|


|
|
PB0040.1_Lef1_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.63
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----ATCAAWTGGA--
NANAGATCAAAGGGNNN |
|


|
|
MA0058.2_MAX/Jaspar
| Match Rank: | 3 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -ATCAAWTGGA
AAGCACATGG- |
|


|
|
PB0083.1_Tcf7_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.62
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----ATCAAWTGGA--
TATAGATCAAAGGAAAA |
|


|
|
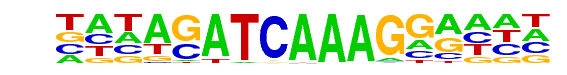
PB0084.1_Tcf7l2_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.62
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----ATCAAWTGGA--
NNNAGATCAAAGGANNN |
|

|
|
Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer
| Match Rank: | 6 |
| Score: | 0.62
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ATCAAWTGGA
ACATCAAAGGNA |
|


|
|
MA0147.2_Myc/Jaspar
| Match Rank: | 7 |
| Score: | 0.62
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ATCAAWTGGA
AAGCACATGG- |
|


|
|
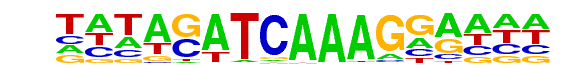
PB0082.1_Tcf3_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----ATCAAWTGGA--
TATAGATCAAAGGAAAA |
|

|
|
Olig2(bHLH)/Neuron-Olig2-ChIP-Seq(GSE30882)/Homer
| Match Rank: | 9 |
| Score: | 0.61
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATCAAWTGGA
AACAKATGGY |
|


|
|
Usf2(HLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer
| Match Rank: | 10 |
| Score: | 0.61
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ATCAAWTGGA
GTCACGTGGT |
|


|
|





















