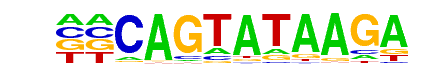
MA0108.2_TBP/Jaspar
| Match Rank: | 1 |
| Score: | 0.60
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CAGTATAAGA-------
--GTATAAAAGGCGGGG |
|


|
|
PB0174.1_Sox30_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.59
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CAGTATAAGA---
TAAGATTATAATACGG |
|


|
|
POL012.1_TATA-Box/Jaspar
| Match Rank: | 3 |
| Score: | 0.59
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CAGTATAAGA-------
--GTATAAAAGGCGGGG |
|


|
|
PB0079.1_Sry_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.58
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CAGTATAAGA---
TATAATTATAATATTC |
|


|
|
MA0032.1_FOXC1/Jaspar
| Match Rank: | 5 |
| Score: | 0.58
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CAGTATAAGA
GGTAAGTA----- |
|


|
|
PB0206.1_Zic2_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.55
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CAGTATAAGA
CCACACAGCAGGAGA |
|


|
|
POL009.1_DCE_S_II/Jaspar
| Match Rank: | 7 |
| Score: | 0.55
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CAGTATAAGA
CACAGN------ |
|

|
|
POL002.1_INR/Jaspar
| Match Rank: | 8 |
| Score: | 0.55
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CAGTATAAGA
TCAGTCTT--- |
|


|
|
PB0105.1_Arid3a_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.55
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CAGTATAAGA---
ACCCGTATCAAATTT |
|


|
|
PB0069.1_Sox21_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.54
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CAGTATAAGA---
TTTAATTATAATTAAG |
|


|
|





















