IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer
| Match Rank: | 1 |
| Score: | 0.53
| | Offset: | 7
| | Orientation: | reverse strand |
| Alignment: | CSCYGBBGCTKTYKGCYSCV
-------RSTTTCRSTTTC- |
|


|
|
MA0528.1_ZNF263/Jaspar
| Match Rank: | 2 |
| Score: | 0.49
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CSCYGBBGCTKTYKGCYSCV
TCCTCCTCCCCCTCCTCCTCC- |
|


|
|
MA0050.2_IRF1/Jaspar
| Match Rank: | 3 |
| Score: | 0.49
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CSCYGBBGCTKTYKGCYSCV
TTTTACTTTCACTTTCACTTT-- |
|


|
|
IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer
| Match Rank: | 4 |
| Score: | 0.49
| | Offset: | 7
| | Orientation: | reverse strand |
| Alignment: | CSCYGBBGCTKTYKGCYSCV
-------ACTTTCACTTTC- |
|


|
|
MA0514.1_Sox3/Jaspar
| Match Rank: | 5 |
| Score: | 0.49
| | Offset: | 7
| | Orientation: | forward strand |
| Alignment: | CSCYGBBGCTKTYKGCYSCV
-------CCTTTGTTTT--- |
|


|
|
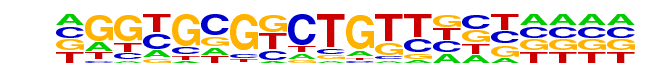
PB0151.1_Myf6_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.48
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CSCYGBBGCTKTYKGCYSCV
-GGNGCGNCTGTTNNN---- |
|

|
|
MA0051.1_IRF2/Jaspar
| Match Rank: | 7 |
| Score: | 0.47
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----CSCYGBBGCTKTYKGCYSCV
GTTTTGCTTTCACTTTCC------ |
|


|
|
ISRE(IRF)/ThioMac-LPS-exp(GSE23622)/HOMER
| Match Rank: | 8 |
| Score: | 0.47
| | Offset: | 7
| | Orientation: | forward strand |
| Alignment: | CSCYGBBGCTKTYKGCYSCV
-------AGTTTCAGTTTC- |
|


|
|
MA0517.1_STAT2::STAT1/Jaspar
| Match Rank: | 9 |
| Score: | 0.47
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | CSCYGBBGCTKTYKGCYSCV
-----TCAGTTTCATTTTCC |
|


|
|
MA0508.1_PRDM1/Jaspar
| Match Rank: | 10 |
| Score: | 0.47
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CSCYGBBGCTKTYKGCYSCV
TCACTTTCACTTTCN------ |
|


|
|





















