MA0524.1_TFAP2C/Jaspar
| Match Rank: | 1 |
| Score: | 0.51
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CAGMGCCCGGCSGSYGCKCC
---TGCCCTGGGGCNANN-- |
|


|
|
BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 2 |
| Score: | 0.47
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CAGMGCCCGGCSGSYGCKCC
CNNBRGCGCCCCCTGSTGGC----- |
|


|
|
MA0003.2_TFAP2A/Jaspar
| Match Rank: | 3 |
| Score: | 0.46
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CAGMGCCCGGCSGSYGCKCC
---TGCCCTGAGGCANTN-- |
|


|
|
POL013.1_MED-1/Jaspar
| Match Rank: | 4 |
| Score: | 0.46
| | Offset: | 15
| | Orientation: | forward strand |
| Alignment: | CAGMGCCCGGCSGSYGCKCC-
---------------GCTCCG |
|


|
|
PB0003.1_Ascl2_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.46
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | CAGMGCCCGGCSGSYGCKCC--
-----CTCAGCAGCTGCTACTG |
|


|
|
MA0154.2_EBF1/Jaspar
| Match Rank: | 6 |
| Score: | 0.45
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CAGMGCCCGGCSGSYGCKCC
----TCCCTGGGGAN----- |
|


|
|
Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 7 |
| Score: | 0.45
| | Offset: | 11
| | Orientation: | forward strand |
| Alignment: | CAGMGCCCGGCSGSYGCKCC-
-----------CNGTCCTCCC |
|


|
|
AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 8 |
| Score: | 0.44
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CAGMGCCCGGCSGSYGCKCC
--ATGCCCTGAGGC------ |
|


|
|
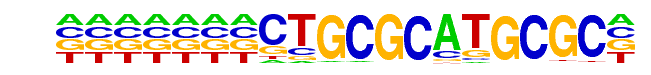
NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 9 |
| Score: | 0.44
| | Offset: | 7
| | Orientation: | forward strand |
| Alignment: | CAGMGCCCGGCSGSYGCKCC
-------CTGCGCATGCGC- |
|

|
|
AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer
| Match Rank: | 10 |
| Score: | 0.44
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CAGMGCCCGGCSGSYGCKCC
--WTGSCCTSAGGS------ |
|


|
|





















