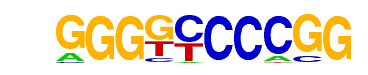
PB0052.1_Plagl1_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.70
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GGGGCCCCGG--
TTGGGGGCGCCCCTAG |
|


|
|
PB0057.1_Rxra_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.56
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----GGGGCCCCGG--
NTNNNGGGGTCANGNNN |
|


|
|
MA0163.1_PLAG1/Jaspar
| Match Rank: | 3 |
| Score: | 0.55
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GGGGCCCCGG----
GGGGCCCAAGGGGG |
|


|
|
POL011.1_XCPE1/Jaspar
| Match Rank: | 4 |
| Score: | 0.55
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GGGGCCCCGG--
--GGTCCCGCCC |
|


|
|
MA0056.1_MZF1_1-4/Jaspar
| Match Rank: | 5 |
| Score: | 0.55
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | GGGGCCCCGG
---TCCCCA- |
|
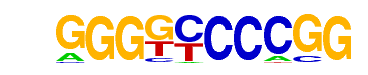

|
|
EBF(EBF)/proBcell-EBF-ChIP-Seq(GSE21978)/Homer
| Match Rank: | 6 |
| Score: | 0.54
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GGGGCCCCGG---
-NGTCCCNNGGGA |
|


|
|
EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 7 |
| Score: | 0.54
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | GGGGCCCCGG----
--GTCCCCAGGGGA |
|


|
|
PB0025.1_Glis2_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.54
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------GGGGCCCCGG
NTNTGGGGGGTCNNNA |
|


|
|
Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 9 |
| Score: | 0.53
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | GGGGCCCCGG---
---TTCCNGGAAG |
|


|
|
PB0156.1_Plagl1_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.53
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----GGGGCCCCGG--
GCTGGGGGGTACCCCTT |
|


|
|