MA0524.1_TFAP2C/Jaspar
| Match Rank: | 1 |
| Score: | 0.69
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CCTGAAGGAA--
TGCCCTGGGGCNANN |
|


|
|
POL008.1_DCE_S_I/Jaspar
| Match Rank: | 2 |
| Score: | 0.66
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CCTGAAGGAA
--NGAAGC-- |
|


|
|
MA0028.1_ELK1/Jaspar
| Match Rank: | 3 |
| Score: | 0.63
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CCTGAAGGAA
GAGCCGGAAG--- |
|


|
|
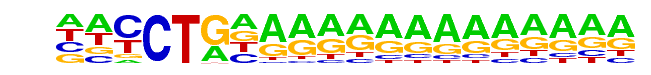
SA0002.1_at_AC_acceptor/Jaspar
| Match Rank: | 4 |
| Score: | 0.63
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CCTGAAGGAA--------
NNCCTGNAAAAAAAAAAAAA |
|

|
|
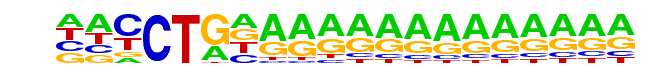
SA0001.1_at_AC_acceptor/Jaspar
| Match Rank: | 5 |
| Score: | 0.62
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CCTGAAGGAA--------
NNCCTGNAAAAAAAAAAAAA |
|

|
|
AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer
| Match Rank: | 6 |
| Score: | 0.61
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CCTGAAGGAA-
SCCTSAGGSCAW |
|


|
|
Unknown-ESC-element/mES-Nanog-ChIP-Seq(GSE11724)/Homer
| Match Rank: | 7 |
| Score: | 0.61
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CCTGAAGGAA-
CACAGCAGGGGG |
|


|
|
MF0001.1_ETS_class/Jaspar
| Match Rank: | 8 |
| Score: | 0.61
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CCTGAAGGAA
ACCGGAAG--- |
|


|
|
ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 9 |
| Score: | 0.61
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CCTGAAGGAA
ANCCGGAAGT-- |
|


|
|
PB0205.1_Zic1_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.60
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CCTGAAGGAA-
CCACACAGCAGGAGA |
|


|
|





















