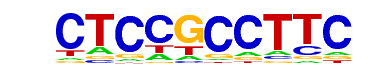
MA0079.3_SP1/Jaspar
| Match Rank: | 1 |
| Score: | 0.70
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CTCCGCCTTC
GCCCCGCCCCC |
|


|
|
POL003.1_GC-box/Jaspar
| Match Rank: | 2 |
| Score: | 0.68
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CTCCGCCTTC-
NAGCCCCGCCCCCN |
|


|
|
MA0516.1_SP2/Jaspar
| Match Rank: | 3 |
| Score: | 0.68
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CTCCGCCTTC----
GCCCCGCCCCCTCCC |
|


|
|
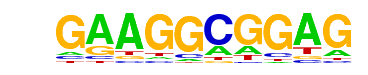
MA0162.2_EGR1/Jaspar
| Match Rank: | 4 |
| Score: | 0.64
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CTCCGCCTTC---
CCCCCGCCCCCGCC |
|

|
|
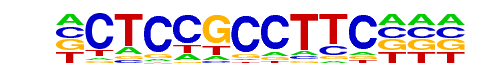
MA0057.1_MZF1_5-13/Jaspar
| Match Rank: | 5 |
| Score: | 0.62
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CTCCGCCTTC
TTCCCCCTAC |
|

|
|
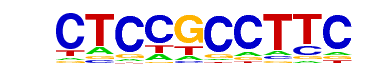
Sp1(Zf)/Promoter/Homer
| Match Rank: | 6 |
| Score: | 0.62
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CTCCGCCTTC
GGCCCCGCCCCC |
|


|
|
POL008.1_DCE_S_I/Jaspar
| Match Rank: | 7 |
| Score: | 0.61
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | CTCCGCCTTC-
-----GCTTCC |
|


|
|
MA0528.1_ZNF263/Jaspar
| Match Rank: | 8 |
| Score: | 0.60
| | Offset: | -11
| | Orientation: | reverse strand |
| Alignment: | -----------CTCCGCCTTC
TCCTCCTCCCCCTCCTCCTCC |
|


|
|
PB0164.1_Smad3_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.60
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CTCCGCCTTC---
TACGCCCCGCCACTCTG |
|


|
|
PB0076.1_Sp4_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.59
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CTCCGCCTTC--
GGTCCCGCCCCCTTCTC |
|


|
|