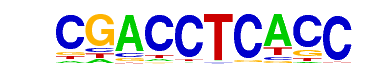
MA0103.2_ZEB1/Jaspar
| Match Rank: | 1 |
| Score: | 0.65
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CGACCTCACC--
---CCTCACCTG |
|


|
|
MA0493.1_Klf1/Jaspar
| Match Rank: | 2 |
| Score: | 0.64
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CGACCTCACC--
-GGCCACACCCA |
|


|
|
EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer
| Match Rank: | 3 |
| Score: | 0.64
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CGACCTCACC---
-GGCCACACCCAN |
|


|
|
PB0039.1_Klf7_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CGACCTCACC-----
TCGACCCCGCCCCTAT |
|


|
|
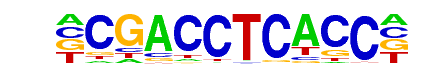
c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 5 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CGACCTCACC-
ATGACGTCATCN |
|

|
|
MA0039.2_Klf4/Jaspar
| Match Rank: | 6 |
| Score: | 0.61
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CGACCTCACC--
--GCCCCACCCA |
|


|
|
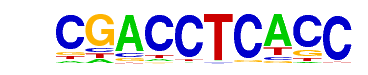
MA0018.2_CREB1/Jaspar
| Match Rank: | 7 |
| Score: | 0.61
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CGACCTCACC
TGACGTCA-- |
|

|
|
SD0001.1_at_AC_acceptor/Jaspar
| Match Rank: | 8 |
| Score: | 0.61
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CGACCTCACC--
-NNACTTACCTN |
|


|
|
MA0492.1_JUND_(var.2)/Jaspar
| Match Rank: | 9 |
| Score: | 0.60
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CGACCTCACC---
NATGACATCATCNNN |
|


|
|
KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer
| Match Rank: | 10 |
| Score: | 0.60
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CGACCTCACC--
--GCCMCRCCCH |
|


|
|