





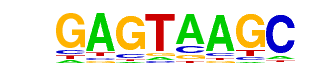


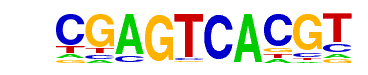





| p-value: | 1e-960 |
| log p-value: | -2.212e+03 |
| Information Content per bp: | 1.587 |
| Number of Target Sequences with motif | 1613.0 |
| Percentage of Target Sequences with motif | 36.93% |
| Number of Background Sequences with motif | 163.4 |
| Percentage of Background Sequences with motif | 4.65% |
| Average Position of motif in Targets | 150.2 +/- 60.3bp |
| Average Position of motif in Background | 141.4 +/- 93.3bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.15 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.981 |  | 1e-932 | -2146.888031 | 35.05% | 4.23% | motif file (matrix) |
| 2 | 0.960 |  | 1e-927 | -2135.533161 | 43.27% | 7.37% | motif file (matrix) |
| 3 | 0.933 |  | 1e-612 | -1411.238230 | 30.22% | 5.14% | motif file (matrix) |
| 4 | 0.826 |  | 1e-217 | -500.119375 | 9.13% | 1.15% | motif file (matrix) |
| 5 | 0.838 |  | 1e-166 | -384.193167 | 13.58% | 3.56% | motif file (matrix) |
| 6 | 0.850 |  | 1e-165 | -380.128106 | 8.47% | 1.39% | motif file (matrix) |
| 7 | 0.615 |  | 1e-115 | -266.889265 | 32.55% | 18.11% | motif file (matrix) |
| 8 | 0.774 |  | 1e-95 | -220.472057 | 2.17% | 0.09% | motif file (matrix) |
| 9 | 0.611 |  | 1e-56 | -129.000524 | 3.87% | 0.86% | motif file (matrix) |
| 10 | 0.658 |  | 1e-34 | -79.308943 | 0.94% | 0.08% | motif file (matrix) |
| 11 | 0.643 |  | 1e-33 | -76.681158 | 1.81% | 0.33% | motif file (matrix) |
| 12 | 0.613 |  | 1e-28 | -66.422521 | 3.96% | 1.50% | motif file (matrix) |
| 13 | 0.615 |  | 1e-14 | -34.327414 | 0.80% | 0.15% | motif file (matrix) |