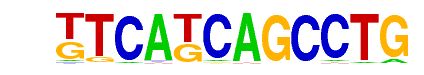
| p-value: | 1e-30 |
| log p-value: | -7.094e+01 |
| Information Content per bp: | 1.845 |
| Number of Target Sequences with motif | 38.0 |
| Percentage of Target Sequences with motif | 0.87% |
| Number of Background Sequences with motif | 2.5 |
| Percentage of Background Sequences with motif | 0.07% |
| Average Position of motif in Targets | 138.9 +/- 81.8bp |
| Average Position of motif in Background | 114.9 +/- 23.2bp |
| Strand Bias (log2 ratio + to - strand density) | -0.6 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0117.1_Mafb/Jaspar
| Match Rank: | 1 |
| Score: | 0.62
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CAGGCTGATGAA
---GCTGACGC- |
|


|
|
POL010.1_DCE_S_III/Jaspar
| Match Rank: | 2 |
| Score: | 0.60
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CAGGCTGATGAA
--NGCTN----- |
|


|
|
Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer
| Match Rank: | 3 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CAGGCTGATGAA
TWGTCTGV---- |
|


|
|
Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer
| Match Rank: | 4 |
| Score: | 0.59
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CAGGCTGATGAA---
---GGTGYTGACAGS |
|


|
|
SCL(HLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer
| Match Rank: | 5 |
| Score: | 0.58
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CAGGCTGATGAA
--ANCAGCTG-- |
|


|
|
MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer
| Match Rank: | 6 |
| Score: | 0.58
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | CAGGCTGATGAA---
-----TGCTGACTCA |
|


|
|
Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 7 |
| Score: | 0.56
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CAGGCTGATGAA--
----ATGATGCAAT |
|


|
|
MA0496.1_MAFK/Jaspar
| Match Rank: | 8 |
| Score: | 0.56
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CAGGCTGATGAA----
-AAANTGCTGACTNAG |
|


|
|
Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 9 |
| Score: | 0.55
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | CAGGCTGATGAA--
----MTGATGCAAT |
|


|
|
PH0016.1_Cux1_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.53
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CAGGCTGATGAA----
ACCGGTTGATCACCTGA |
|


|
|