| p-value: | 1e-30 |
| log p-value: | -6.944e+01 |
| Information Content per bp: | 1.854 |
| Number of Target Sequences with motif | 30.0 |
| Percentage of Target Sequences with motif | 0.69% |
| Number of Background Sequences with motif | 0.7 |
| Percentage of Background Sequences with motif | 0.02% |
| Average Position of motif in Targets | 175.6 +/- 88.2bp |
| Average Position of motif in Background | 196.0 +/- 0.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.3 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
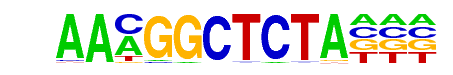
PRDM14(Zf)/H1-PRDM14-ChIP-Seq(GSE22767)/Homer
| Match Rank: | 1 |
| Score: | 0.63
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AAMGGCTCTA---
-AGGTCTCTAACC |
|
|
|
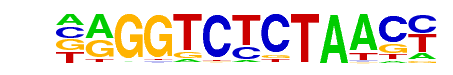
POL010.1_DCE_S_III/Jaspar
| Match Rank: | 2 |
| Score: | 0.61
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | AAMGGCTCTA
---NGCTN-- |
|


|
|
BMYB(HTH)/Hela-BMYB-ChIPSeq(GSE27030)/Homer
| Match Rank: | 3 |
| Score: | 0.59
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AAMGGCTCTA
NHAACBGYYV-- |
|


|
|
PB0194.1_Zbtb12_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.58
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AAMGGCTCTA------
-AGNGTTCTAATGANN |
|


|
|
PB0046.1_Mybl1_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.56
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----AAMGGCTCTA--
NNANTAACGGTTNNNAN |
|


|
|
PB0090.1_Zbtb12_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.55
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AAMGGCTCTA------
CTAAGGTTCTAGATCAC |
|


|
|
PB0045.1_Myb_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.54
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----AAMGGCTCTA--
NNNNTAACGGTTNNNAN |
|


|
|
PB0137.1_Irf3_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.54
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----AAMGGCTCTA
GGAGAAAGGTGCGA |
|


|
|
MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer
| Match Rank: | 9 |
| Score: | 0.54
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AAMGGCTCTA
YAACBGCC--- |
|


|
|
PB0113.1_E2F3_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.52
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AAMGGCTCTA-----
AGCTCGGCGCCAAAAGC |
|


|
|





















