| p-value: | 1e-51 |
| log p-value: | -1.188e+02 |
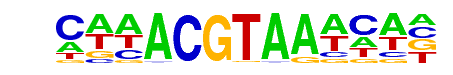
| Information Content per bp: | 1.642 |
| Number of Target Sequences with motif | 298.0 |
| Percentage of Target Sequences with motif | 30.32% |
| Number of Background Sequences with motif | 53.7 |
| Percentage of Background Sequences with motif | 12.20% |
| Average Position of motif in Targets | 160.6 +/- 86.7bp |
| Average Position of motif in Background | 146.7 +/- 68.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.21 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0027.1_Gmeb1_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.78
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----CWWACGTAAWCA-
NNNTNGTACGTAANNNN |
|


|
|
HIF-1a(HLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer
| Match Rank: | 2 |
| Score: | 0.75
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CWWACGTAAWCA
BGCACGTA---- |
|


|
|
MA0030.1_FOXF2/Jaspar
| Match Rank: | 3 |
| Score: | 0.75
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CWWACGTAAWCA--
CAAACGTAAACAAT |
|


|
|
MF0002.1_bZIP_CREB/G-box-like_subclass/Jaspar
| Match Rank: | 4 |
| Score: | 0.73
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CWWACGTAAWCA
---ACGTCA--- |
|


|
|
MA0025.1_NFIL3/Jaspar
| Match Rank: | 5 |
| Score: | 0.72
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CWWACGTAAWCA
-TTATGTAACAT |
|


|
|
HIF2a(HLH)/785_O-HIF2a-ChIP-Seq(GSE34871)/Homer
| Match Rank: | 6 |
| Score: | 0.70
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CWWACGTAAWCA
-GCACGTACCC- |
|


|
|
MA0031.1_FOXD1/Jaspar
| Match Rank: | 7 |
| Score: | 0.69
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | CWWACGTAAWCA-
-----GTAAACAT |
|

|
|
FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer
| Match Rank: | 8 |
| Score: | 0.68
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CWWACGTAAWCA
--AAAGTAAACA |
|


|
|
MA0043.1_HLF/Jaspar
| Match Rank: | 9 |
| Score: | 0.68
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CWWACGTAAWCA
NATTACGTAACC- |
|


|
|
FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer
| Match Rank: | 10 |
| Score: | 0.67
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CWWACGTAAWCA
--AAAGTAAACA |
|


|
|





















