| p-value: | 1e-98 |
| log p-value: | -2.258e+02 |
| Information Content per bp: | 1.814 |
| Number of Target Sequences with motif | 83.0 |
| Percentage of Target Sequences with motif | 8.44% |
| Number of Background Sequences with motif | 1.5 |
| Percentage of Background Sequences with motif | 0.34% |
| Average Position of motif in Targets | 158.5 +/- 83.2bp |
| Average Position of motif in Background | 180.3 +/- 58.8bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.05 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
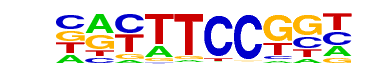
Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer
| Match Rank: | 1 |
| Score: | 0.79
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGATTCCG--
NRYTTCCGGH |
|

|
|
Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 2 |
| Score: | 0.78
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGATTCCG--
NRYTTCCGGY |
|


|
|
Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 3 |
| Score: | 0.76
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGATTCCG--
HACTTCCGGY |
|


|
|
MA0081.1_SPIB/Jaspar
| Match Rank: | 4 |
| Score: | 0.75
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CGATTCCG--
---TTCCTCT |
|


|
|
MA0028.1_ELK1/Jaspar
| Match Rank: | 5 |
| Score: | 0.74
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CGATTCCG----
--CTTCCGGNNN |
|


|
|
MF0001.1_ETS_class/Jaspar
| Match Rank: | 6 |
| Score: | 0.73
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CGATTCCG--
--CTTCCGGT |
|


|
|
MA0076.2_ELK4/Jaspar
| Match Rank: | 7 |
| Score: | 0.72
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CGATTCCG--
CCACTTCCGGC |
|


|
|
MA0062.2_GABPA/Jaspar
| Match Rank: | 8 |
| Score: | 0.71
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CGATTCCG-
NCCACTTCCGG |
|


|
|
TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer
| Match Rank: | 9 |
| Score: | 0.69
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CGATTCCG--
RCATTCCWGG |
|


|
|
ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 10 |
| Score: | 0.68
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CGATTCCG---
-ACTTCCGGNT |
|


|
|





















