





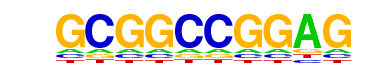


| p-value: | 1e-432 |
| log p-value: | -9.966e+02 |
| Information Content per bp: | 1.357 |
| Number of Target Sequences with motif | 276.0 |
| Percentage of Target Sequences with motif | 12.31% |
| Number of Background Sequences with motif | 1.8 |
| Percentage of Background Sequences with motif | 0.25% |
| Average Position of motif in Targets | 142.0 +/- 80.4bp |
| Average Position of motif in Background | 188.6 +/- 53.1bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.14 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.637 |  | 1e-397 | -916.368720 | 13.51% | 0.34% | motif file (matrix) |
| 2 | 0.624 |  | 1e-278 | -641.619354 | 10.39% | 0.35% | motif file (matrix) |
| 3 | 0.663 |  | 1e-249 | -574.541605 | 9.59% | 0.37% | motif file (matrix) |
| 4 | 0.819 |  | 1e-248 | -572.350957 | 13.87% | 0.93% | motif file (matrix) |
| 5 | 0.638 |  | 1e-218 | -503.208625 | 9.77% | 0.45% | motif file (matrix) |
| 6 | 0.771 |  | 1e-190 | -439.024795 | 10.44% | 0.68% | motif file (matrix) |
| 7 | 0.621 |  | 1e-84 | -193.604811 | 6.69% | 0.83% | motif file (matrix) |