






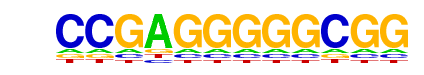



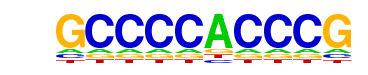



| p-value: | 1e-406 |
| log p-value: | -9.365e+02 |
| Information Content per bp: | 1.618 |
| Number of Target Sequences with motif | 263.0 |
| Percentage of Target Sequences with motif | 11.73% |
| Number of Background Sequences with motif | 1.6 |
| Percentage of Background Sequences with motif | 0.21% |
| Average Position of motif in Targets | 157.3 +/- 80.3bp |
| Average Position of motif in Background | 130.7 +/- 53.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.21 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.655 |  | 1e-357 | -823.830666 | 12.49% | 0.38% | motif file (matrix) |
| 2 | 0.615 |  | 1e-316 | -729.571701 | 11.42% | 0.39% | motif file (matrix) |
| 3 | 0.625 |  | 1e-305 | -703.407170 | 9.41% | 0.00% | motif file (matrix) |
| 4 | 0.746 |  | 1e-229 | -528.986510 | 11.02% | 0.61% | motif file (matrix) |
| 5 | 0.616 |  | 1e-222 | -513.116477 | 9.90% | 0.48% | motif file (matrix) |
| 6 | 0.650 |  | 1e-218 | -503.208625 | 9.77% | 0.47% | motif file (matrix) |
| 7 | 0.786 |  | 1e-213 | -491.271316 | 8.56% | 0.36% | motif file (matrix) |
| 8 | 0.622 |  | 1e-191 | -442.037927 | 7.94% | 0.30% | motif file (matrix) |
| 9 | 0.742 |  | 1e-169 | -390.614836 | 7.27% | 0.35% | motif file (matrix) |
| 10 | 0.663 |  | 1e-132 | -305.079941 | 6.11% | 0.37% | motif file (matrix) |
| 11 | 0.632 |  | 1e-126 | -291.354717 | 13.83% | 2.66% | motif file (matrix) |
| 12 | 0.702 |  | 1e-95 | -219.447342 | 5.58% | 0.45% | motif file (matrix) |
| 13 | 0.622 |  | 1e-79 | -183.238053 | 8.07% | 1.46% | motif file (matrix) |