








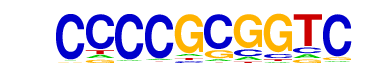
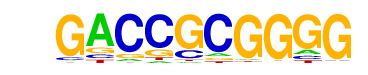
| p-value: | 1e-104 |
| log p-value: | -2.415e+02 |
| Information Content per bp: | 1.665 |
| Number of Target Sequences with motif | 76.0 |
| Percentage of Target Sequences with motif | 18.81% |
| Number of Background Sequences with motif | 2.0 |
| Percentage of Background Sequences with motif | 0.66% |
| Average Position of motif in Targets | 927.2 +/- 992.5bp |
| Average Position of motif in Background | 2019.9 +/- 756.3bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.29 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.791 |  | 1e-59 | -135.957593 | 22.52% | 2.66% | motif file (matrix) |
| 2 | 0.646 |  | 1e-54 | -126.305000 | 11.63% | 0.57% | motif file (matrix) |
| 3 | 0.608 |  | 1e-43 | -101.207445 | 9.90% | 0.25% | motif file (matrix) |
| 4 | 0.620 |  | 1e-40 | -92.861510 | 13.12% | 1.11% | motif file (matrix) |
| 5 | 0.628 |  | 1e-37 | -87.457320 | 8.91% | 0.65% | motif file (matrix) |
| 6 | 0.629 |  | 1e-21 | -48.893025 | 17.08% | 4.46% | motif file (matrix) |
| 7 | 0.748 |  | 1e-19 | -45.219929 | 9.41% | 1.68% | motif file (matrix) |
| 8 | 0.626 |  | 1e-16 | -37.322662 | 13.12% | 3.66% | motif file (matrix) |
| 9 | 0.632 |  | 1e-11 | -27.603229 | 9.41% | 2.65% | motif file (matrix) |