| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | HNF6(Homeobox)/Liver-Hnf6-ChIP-Seq(ERP000394)/Homer | 1e-15 | -3.628e+01 | 0.0000 | 70.0 | 17.33% | 17.9 | 6.03% | motif file (matrix) |
pdf |
| 2 |  | PRDM9(Zf)/Testis-DMC1-ChIP-Seq(GSE35498)/Homer | 1e-12 | -2.828e+01 | 0.0000 | 72.0 | 17.82% | 21.9 | 7.36% | motif file (matrix) |
pdf |
| 3 |  | ETS(ETS)/Promoter/Homer | 1e-10 | -2.325e+01 | 0.0000 | 79.0 | 19.55% | 27.7 | 9.30% | motif file (matrix) |
pdf |
| 4 |  | GFY-Staf/Promoters/Homer | 1e-8 | -1.997e+01 | 0.0000 | 13.0 | 3.22% | 1.3 | 0.42% | motif file (matrix) |
pdf |
| 5 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-8 | -1.885e+01 | 0.0000 | 111.0 | 27.48% | 48.6 | 16.33% | motif file (matrix) |
pdf |
| 6 |  | RBPJ:Ebox/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-8 | -1.853e+01 | 0.0000 | 56.0 | 13.86% | 18.8 | 6.34% | motif file (matrix) |
pdf |
| 7 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-7 | -1.760e+01 | 0.0000 | 57.0 | 14.11% | 19.3 | 6.49% | motif file (matrix) |
pdf |
| 8 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-7 | -1.684e+01 | 0.0000 | 62.0 | 15.35% | 22.8 | 7.66% | motif file (matrix) |
pdf |
| 9 |  | PAX5-shortForm(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-7 | -1.674e+01 | 0.0000 | 19.0 | 4.70% | 3.4 | 1.13% | motif file (matrix) |
pdf |
| 10 |  | TR4(NR/DR1)/Hela-TR4-ChIP-Seq(GSE24685)/Homer | 1e-7 | -1.674e+01 | 0.0000 | 19.0 | 4.70% | 4.0 | 1.33% | motif file (matrix) |
pdf |
| 11 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-6 | -1.420e+01 | 0.0000 | 82.0 | 20.30% | 35.3 | 11.85% | motif file (matrix) |
pdf |
| 12 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-6 | -1.396e+01 | 0.0000 | 30.0 | 7.43% | 8.2 | 2.75% | motif file (matrix) |
pdf |
| 13 |  | GFY(?)/Promoter/Homer | 1e-5 | -1.334e+01 | 0.0000 | 10.0 | 2.48% | 1.3 | 0.43% | motif file (matrix) |
pdf |
| 14 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-5 | -1.333e+01 | 0.0000 | 72.0 | 17.82% | 30.8 | 10.34% | motif file (matrix) |
pdf |
| 15 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-5 | -1.307e+01 | 0.0000 | 135.0 | 33.42% | 69.8 | 23.47% | motif file (matrix) |
pdf |
| 16 |  | Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer | 1e-5 | -1.266e+01 | 0.0000 | 31.0 | 7.67% | 9.7 | 3.26% | motif file (matrix) |
pdf |
| 17 |  | GRE(NR/IR3)/A549-GR-ChIP-Seq(GSE32465)/Homer | 1e-5 | -1.248e+01 | 0.0001 | 24.0 | 5.94% | 6.2 | 2.09% | motif file (matrix) |
pdf |
| 18 |  | GRE/RAW264.7-GRE-ChIP-Seq(Unpublished)/Homer | 1e-5 | -1.233e+01 | 0.0001 | 37.0 | 9.16% | 12.6 | 4.23% | motif file (matrix) |
pdf |
| 19 |  | TRa(NR)/C17.2-TRa-ChIP-Seq(GSE38347)/Homer | 1e-5 | -1.233e+01 | 0.0001 | 67.0 | 16.58% | 29.0 | 9.74% | motif file (matrix) |
pdf |
| 20 |  | Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo et al.)/Homer | 1e-5 | -1.232e+01 | 0.0001 | 107.0 | 26.49% | 52.5 | 17.66% | motif file (matrix) |
pdf |
| 21 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-5 | -1.209e+01 | 0.0001 | 77.0 | 19.06% | 34.2 | 11.50% | motif file (matrix) |
pdf |
| 22 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-4 | -1.136e+01 | 0.0001 | 94.0 | 23.27% | 45.3 | 15.24% | motif file (matrix) |
pdf |
| 23 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-4 | -1.130e+01 | 0.0001 | 89.0 | 22.03% | 42.1 | 14.14% | motif file (matrix) |
pdf |
| 24 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-4 | -1.078e+01 | 0.0002 | 43.0 | 10.64% | 16.7 | 5.62% | motif file (matrix) |
pdf |
| 25 |  | Nkx2.5(Homeobox)/HL1-Nkx2.5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-4 | -9.832e+00 | 0.0005 | 223.0 | 55.20% | 135.2 | 45.44% | motif file (matrix) |
pdf |
| 26 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-4 | -9.664e+00 | 0.0006 | 137.0 | 33.91% | 75.3 | 25.29% | motif file (matrix) |
pdf |
| 27 |  | BMYB(HTH)/Hela-BMYB-ChIPSeq(GSE27030)/Homer | 1e-4 | -9.657e+00 | 0.0006 | 162.0 | 40.10% | 92.6 | 31.14% | motif file (matrix) |
pdf |
| 28 |  | FXR(NR/IR1)/Liver-FXR-ChIP-Seq(Chong et al.)/Homer | 1e-4 | -9.369e+00 | 0.0007 | 64.0 | 15.84% | 29.6 | 9.93% | motif file (matrix) |
pdf |
| 29 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-3 | -8.886e+00 | 0.0012 | 27.0 | 6.68% | 9.9 | 3.34% | motif file (matrix) |
pdf |
| 30 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-3 | -8.612e+00 | 0.0015 | 115.0 | 28.47% | 62.1 | 20.88% | motif file (matrix) |
pdf |
| 31 |  | Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.570e+00 | 0.0015 | 79.0 | 19.55% | 39.6 | 13.31% | motif file (matrix) |
pdf |
| 32 |  | CHR/Cell-Cycle-Exp/Homer | 1e-3 | -8.375e+00 | 0.0018 | 77.0 | 19.06% | 38.8 | 13.03% | motif file (matrix) |
pdf |
| 33 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-3 | -8.373e+00 | 0.0018 | 96.0 | 23.76% | 50.2 | 16.88% | motif file (matrix) |
pdf |
| 34 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-3 | -8.169e+00 | 0.0020 | 50.0 | 12.38% | 22.8 | 7.66% | motif file (matrix) |
pdf |
| 35 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-3 | -8.088e+00 | 0.0022 | 55.0 | 13.61% | 25.7 | 8.64% | motif file (matrix) |
pdf |
| 36 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-3 | -7.931e+00 | 0.0025 | 172.0 | 42.57% | 102.9 | 34.57% | motif file (matrix) |
pdf |
| 37 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.757e+00 | 0.0028 | 119.0 | 29.46% | 66.5 | 22.35% | motif file (matrix) |
pdf |
| 38 | 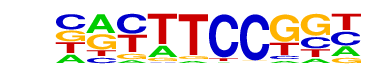 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-3 | -7.719e+00 | 0.0029 | 170.0 | 42.08% | 101.8 | 34.21% | motif file (matrix) |
pdf |
| 39 |  | GATA-DR8(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-3 | -7.600e+00 | 0.0032 | 10.0 | 2.48% | 2.4 | 0.81% | motif file (matrix) |
pdf |
| 40 |  | Mouse_Recombination_Hotspot/Testis-DMC1-ChIP-Seq(GSE24438)/Homer | 1e-3 | -7.583e+00 | 0.0032 | 7.0 | 1.73% | 1.8 | 0.59% | motif file (matrix) |
pdf |
| 41 |  | CEBP(bZIP)/CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.552e+00 | 0.0032 | 64.0 | 15.84% | 31.6 | 10.61% | motif file (matrix) |
pdf |
| 42 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-3 | -7.527e+00 | 0.0032 | 91.0 | 22.52% | 48.8 | 16.42% | motif file (matrix) |
pdf |
| 43 |  | CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.499e+00 | 0.0032 | 72.0 | 17.82% | 36.5 | 12.26% | motif file (matrix) |
pdf |
| 44 |  | AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.470e+00 | 0.0032 | 139.0 | 34.41% | 80.6 | 27.11% | motif file (matrix) |
pdf |
| 45 |  | CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer | 1e-3 | -7.418e+00 | 0.0033 | 218.0 | 53.96% | 136.7 | 45.94% | motif file (matrix) |
pdf |
| 46 |  | Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-3 | -7.194e+00 | 0.0040 | 21.0 | 5.20% | 7.9 | 2.66% | motif file (matrix) |
pdf |
| 47 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.060e+00 | 0.0045 | 79.0 | 19.55% | 41.1 | 13.80% | motif file (matrix) |
pdf |
| 48 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.739e+00 | 0.0061 | 216.0 | 53.47% | 136.7 | 45.95% | motif file (matrix) |
pdf |
| 49 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.730e+00 | 0.0061 | 30.0 | 7.43% | 12.4 | 4.17% | motif file (matrix) |
pdf |
| 50 |  | HIF-1a(HLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer | 1e-2 | -6.621e+00 | 0.0066 | 37.0 | 9.16% | 16.7 | 5.62% | motif file (matrix) |
pdf |
| 51 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-2 | -6.395e+00 | 0.0081 | 105.0 | 25.99% | 59.4 | 19.98% | motif file (matrix) |
pdf |
| 52 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-2 | -6.252e+00 | 0.0091 | 9.0 | 2.23% | 2.5 | 0.82% | motif file (matrix) |
pdf |
| 53 |  | NRF1/Promoter/Homer | 1e-2 | -6.140e+00 | 0.0100 | 53.0 | 13.12% | 26.2 | 8.80% | motif file (matrix) |
pdf |
| 54 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-2 | -6.061e+00 | 0.0106 | 72.0 | 17.82% | 38.1 | 12.80% | motif file (matrix) |
pdf |
| 55 |  | HRE(HSF)/HepG2-HSF1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.711e+00 | 0.0148 | 32.0 | 7.92% | 14.1 | 4.74% | motif file (matrix) |
pdf |
| 56 |  | GATA-IR3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.569e+00 | 0.0168 | 13.0 | 3.22% | 4.2 | 1.42% | motif file (matrix) |
pdf |
| 57 |  | GLI3(Zf)/GLI3-ChIP-Chip(GSE11077)/Homer | 1e-2 | -5.497e+00 | 0.0177 | 17.0 | 4.21% | 6.3 | 2.13% | motif file (matrix) |
pdf |
| 58 |  | Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer | 1e-2 | -5.285e+00 | 0.0215 | 33.0 | 8.17% | 15.3 | 5.13% | motif file (matrix) |
pdf |
| 59 |  | PR(NR)/T47D-PR-ChIP-Seq(GSE31130)/Homer | 1e-2 | -5.069e+00 | 0.0262 | 216.0 | 53.47% | 140.1 | 47.10% | motif file (matrix) |
pdf |
| 60 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.005e+00 | 0.0275 | 8.0 | 1.98% | 2.4 | 0.80% | motif file (matrix) |
pdf |
| 61 | 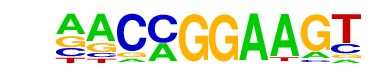 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-2 | -4.933e+00 | 0.0291 | 188.0 | 46.53% | 120.7 | 40.58% | motif file (matrix) |
pdf |
| 62 |  | EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer | 1e-2 | -4.905e+00 | 0.0294 | 39.0 | 9.65% | 19.3 | 6.48% | motif file (matrix) |
pdf |
| 63 |  | Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer | 1e-2 | -4.764e+00 | 0.0333 | 186.0 | 46.04% | 119.9 | 40.30% | motif file (matrix) |
pdf |
| 64 | 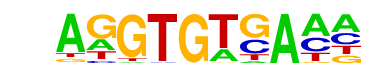 | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-2 | -4.716e+00 | 0.0344 | 109.0 | 26.98% | 65.1 | 21.89% | motif file (matrix) |
pdf |
| 65 |  | HIF2a(HLH)/785_O-HIF2a-ChIP-Seq(GSE34871)/Homer | 1e-2 | -4.714e+00 | 0.0344 | 64.0 | 15.84% | 35.1 | 11.81% | motif file (matrix) |
pdf |
| 66 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-2 | -4.653e+00 | 0.0355 | 25.0 | 6.19% | 11.7 | 3.93% | motif file (matrix) |
pdf |
| 67 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-2 | -4.622e+00 | 0.0361 | 129.0 | 31.93% | 79.7 | 26.78% | motif file (matrix) |
pdf |





































































































































