| p-value: | 1e-15 |
| log p-value: | -3.505e+01 |
| Information Content per bp: | 1.602 |
| Number of Target Sequences with motif | 23.0 |
| Percentage of Target Sequences with motif | 8.30% |
| Number of Background Sequences with motif | 3.4 |
| Percentage of Background Sequences with motif | 0.97% |
| Average Position of motif in Targets | 162.0 +/- 79.1bp |
| Average Position of motif in Background | 160.0 +/- 92.3bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Pax8(Paired/Homeobox)/Thyroid-Pax8-ChIP-Seq(GSE26938)/Homer
| Match Rank: | 1 |
| Score: | 0.67
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AGCCATCCAGGA-
SCAGYCADGCATGAC |
|


|
|
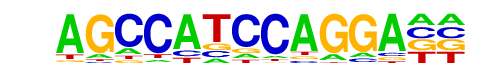
MA0463.1_Bcl6/Jaspar
| Match Rank: | 2 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AGCCATCCAGGA--
NGCTTTCTAGGAAN |
|

|
|
Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer
| Match Rank: | 3 |
| Score: | 0.62
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AGCCATCCAGGA--
NNNCTTTCCAGGAAA |
|


|
|
SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 4 |
| Score: | 0.60
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | AGCCATCCAGGA---
-----ANCAGGATGT |
|


|
|
PAX5(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 5 |
| Score: | 0.60
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AGCCATCCAGGA--
GCAGCCAAGCGTGACN |
|


|
|
Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 6 |
| Score: | 0.58
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | AGCCATCCAGGA-
---CTTCCNGGAA |
|


|
|
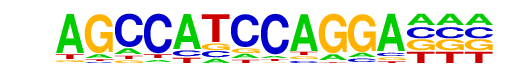
STAT4(Stat)/CD4-Stat4-ChIP-Seq(GSE22104)/Homer
| Match Rank: | 7 |
| Score: | 0.55
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | AGCCATCCAGGA---
---TTTCCNGGAAAN |
|

|
|
POL004.1_CCAAT-box/Jaspar
| Match Rank: | 8 |
| Score: | 0.55
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---AGCCATCCAGGA
ACTAGCCAATCA--- |
|


|
|
ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer
| Match Rank: | 9 |
| Score: | 0.55
| | Offset: | 6
| | Orientation: | forward strand |
| Alignment: | AGCCATCCAGGA----
------ACAGGAAGTG |
|


|
|
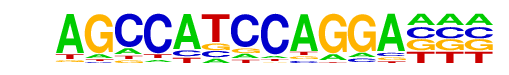
Stat3+il21(Stat)/CD4-Stat3-ChIP-Seq(GSE19198)/Homer
| Match Rank: | 10 |
| Score: | 0.55
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AGCCATCCAGGA---
-NNCTTCCNGGAAGN |
|

|
|





















