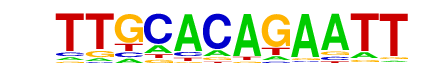
| p-value: | 1e-14 |
| log p-value: | -3.268e+01 |
| Information Content per bp: | 1.604 |
| Number of Target Sequences with motif | 22.0 |
| Percentage of Target Sequences with motif | 7.94% |
| Number of Background Sequences with motif | 3.3 |
| Percentage of Background Sequences with motif | 0.93% |
| Average Position of motif in Targets | 134.0 +/- 88.6bp |
| Average Position of motif in Background | 150.5 +/- 102.9bp |
| Strand Bias (log2 ratio + to - strand density) | 0.8 |
| Multiplicity (# of sites on avg that occur together) | 1.05 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0520.1_Stat6/Jaspar
| Match Rank: | 1 |
| Score: | 0.67
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AATTCTGTGCAA---
ANTTCTCAGGAANNN |
|


|
|
STAT6/Macrophage-Stat6-ChIP-Seq(GSE38377)/Homer
| Match Rank: | 2 |
| Score: | 0.65
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | AATTCTGTGCAA
--TTCTNMGGAA |
|


|
|
PB0208.1_Zscan4_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.64
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AATTCTGTGCAA--
NNNNTTGTGTGCTTNN |
|


|
|
Oct4:Sox17/F9-Sox17-ChIP-Seq(GSE44553)/Homer
| Match Rank: | 4 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AATTCTGTGCAA--
CCATTGTATGCAAAT |
|


|
|
PB0165.1_Sox11_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.61
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AATTCTGTGCAA
AAAATTGTTATGAA |
|


|
|
PB0145.1_Mafb_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AATTCTGTGCAA---
ANATTTTTGCAANTN |
|


|
|
STAT6(Stat)/CD4-Stat6-ChIP-Seq(GSE22104)/Homer
| Match Rank: | 7 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AATTCTGTGCAA
ANTTCTNNAGAA |
|


|
|
PB0176.1_Sox5_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.59
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----AATTCTGTGCAA
NNCTNAATTATGANN-- |
|


|
|
MA0519.1_Stat5a::Stat5b/Jaspar
| Match Rank: | 9 |
| Score: | 0.59
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | AATTCTGTGCAA-
--TTCTTGGAAAN |
|


|
|
CEBP:CEBP(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 10 |
| Score: | 0.59
| | Offset: | -7
| | Orientation: | forward strand |
| Alignment: | -------AATTCTGTGCAA-
NTNATGCAAYMNNHTGMAAY |
|


|
|