| p-value: | 1e-14 |
| log p-value: | -3.402e+01 |
| Information Content per bp: | 1.729 |
| Number of Target Sequences with motif | 32.0 |
| Percentage of Target Sequences with motif | 8.91% |
| Number of Background Sequences with motif | 4.0 |
| Percentage of Background Sequences with motif | 1.50% |
| Average Position of motif in Targets | 153.8 +/- 86.7bp |
| Average Position of motif in Background | 134.5 +/- 103.0bp |
| Strand Bias (log2 ratio + to - strand density) | -1.0 |
| Multiplicity (# of sites on avg that occur together) | 1.20 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0133.1_Hic1_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.76
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GTGCCGTA----
GGGTGTGCCCAAAAGG |
|


|
|
MF0002.1_bZIP_CREB/G-box-like_subclass/Jaspar
| Match Rank: | 2 |
| Score: | 0.62
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GTGCCGTA
-TGACGT- |
|
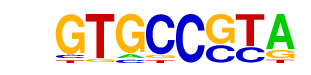

|
|
MA0160.1_NR4A2/Jaspar
| Match Rank: | 3 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GTGCCGTA
GTGACCTT |
|
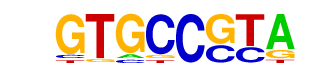

|
|
MA0597.1_THAP1/Jaspar
| Match Rank: | 4 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GTGCCGTA-
CTGCCCGCA |
|


|
|
HIF-1a(HLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer
| Match Rank: | 5 |
| Score: | 0.57
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GTGCCGTA
TACGTGCV--- |
|
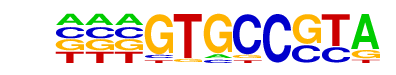

|
|
PB0208.1_Zscan4_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.57
| | Offset: | -8
| | Orientation: | reverse strand |
| Alignment: | --------GTGCCGTA
NNNNTTGTGTGCTTNN |
|


|
|
PB0032.1_IRC900814_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.56
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----GTGCCGTA---
GNNATTTGTCGTAANN |
|
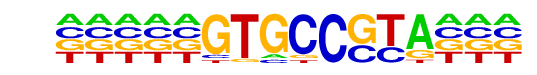

|
|
PB0094.1_Zfp128_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.56
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GTGCCGTA------
TCTTTGGCGTACCCTAA |
|


|
|
PB0130.1_Gm397_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | -8
| | Orientation: | reverse strand |
| Alignment: | --------GTGCCGTA
NNGCGTGTGTGCNGCN |
|


|
|
PB0086.1_Tcfap2b_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GTGCCGTA------
NTGCCCTAGGGCAA |
|


|
|





















