PB0182.1_Srf_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.84
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CAAAAAAA-----
GTTAAAAAAAAAAATTA |
|


|
|
PB0192.1_Tcfap2e_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.84
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CAAAAAAA-
TACTGGAAAAAAAA |
|


|
|
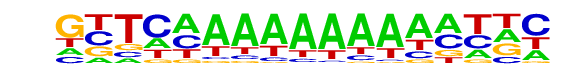
PB0116.1_Elf3_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.80
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CAAAAAAA-----
GTTCAAAAAAAAAATTC |
|

|
|
PH0078.1_Hoxd13/Jaspar
| Match Rank: | 4 |
| Score: | 0.72
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CAAAAAAA----
CTACCAATAAAATTCT |
|


|
|
PB0093.1_Zfp105_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.70
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CAAAAAAA-----
AACAAACAACAAGAG |
|


|
|
PB0145.1_Mafb_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.70
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------CAAAAAAA-
CAATTGCAAAAATAT |
|


|
|
PH0057.1_Hoxb13/Jaspar
| Match Rank: | 7 |
| Score: | 0.70
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CAAAAAAA----
AACCCAATAAAATTCG |
|


|
|
PH0075.1_Hoxd10/Jaspar
| Match Rank: | 8 |
| Score: | 0.69
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CAAAAAAA-----
AATGCAATAAAATTTAT |
|


|
|
PB0186.1_Tcf3_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.69
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CAAAAAAA--
AGCCGAAAAAAAAAT |
|


|
|
Cdx2(Homeobox)/mES-Cdx2-ChIP-Seq(GSE14586)/Homer
| Match Rank: | 10 |
| Score: | 0.69
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CAAAAAAA
GTCATAAAAN |
|


|
|





















