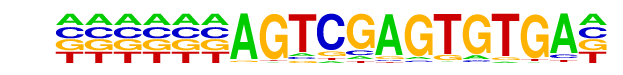
PB0114.1_Egr1_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.61
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGTCGAGTGTGA----
TGCGGAGTGGGACTGG |
|


|
|
PAX5(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 2 |
| Score: | 0.60
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AGTCGAGTGTGA--
GCAGCCAAGCGTGACN |
|


|
|
MA0014.2_PAX5/Jaspar
| Match Rank: | 3 |
| Score: | 0.60
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------AGTCGAGTGTGA-
GAGGGCAGCCAAGCGTGAC |
|

|
|
MA0069.1_Pax6/Jaspar
| Match Rank: | 4 |
| Score: | 0.59
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AGTCGAGTGTGA-
AANTCATGCGTGAA |
|


|
|
Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer
| Match Rank: | 5 |
| Score: | 0.58
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | AGTCGAGTGTGA--
----AGGTGTGAAM |
|


|
|
SD0002.1_at_AC_acceptor/Jaspar
| Match Rank: | 6 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AGTCGAGTGTGA
AAGGCAAGTGT-- |
|


|
|
Nkx2.1(Homeobox)/LungAC-Nkx2.1-ChIP-Seq(GSE43252)/Homer
| Match Rank: | 7 |
| Score: | 0.56
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AGTCGAGTGTGA
-CTYRAGTGSY- |
|


|
|
PB0130.1_Gm397_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AGTCGAGTGTGA----
NNGCGTGTGTGCNGCN |
|


|
|
PB0013.1_Eomes_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGTCGAGTGTGA-----
GAAAAGGTGTGAAAATT |
|


|
|
Nkx2.5(Homeobox)/HL1-Nkx2.5.biotin-ChIP-Seq(GSE21529)/Homer
| Match Rank: | 10 |
| Score: | 0.54
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | AGTCGAGTGTGA
--TTGAGTGSTT |
|


|
|





















