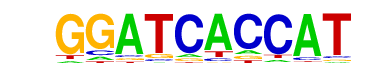
| p-value: | 1e-7 |
| log p-value: | -1.694e+01 |
| Information Content per bp: | 1.651 |
| Number of Target Sequences with motif | 300.0 |
| Percentage of Target Sequences with motif | 3.67% |
| Number of Background Sequences with motif | 206.9 |
| Percentage of Background Sequences with motif | 2.67% |
| Average Position of motif in Targets | 482.7 +/- 441.2bp |
| Average Position of motif in Background | 537.2 +/- 411.4bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.07 |
| Motif File: | file (matrix)
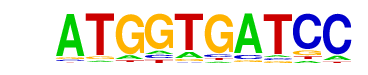
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0196.1_Zbtb7b_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.65
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GGATCACCAT---
CATAAGACCACCATTAC |
|


|
|
PH0017.1_Cux1_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.61
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GGATCACCAT--
TAGTGATCATCATTA |
|


|
|
GATA-IR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer
| Match Rank: | 3 |
| Score: | 0.58
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GGATCACCAT---
NNAGATNVNWATCTN |
|


|
|
Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 4 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GGATCACCAT
GGGAGGACNG- |
|


|
|
CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer
| Match Rank: | 5 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GGATCACCAT
GGATTAGC-- |
|
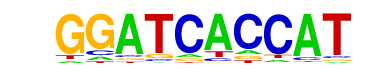

|
|
MA0496.1_MAFK/Jaspar
| Match Rank: | 6 |
| Score: | 0.55
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GGATCACCAT---
CTGAGTCAGCAATTT |
|


|
|
PH0015.1_Crx/Jaspar
| Match Rank: | 7 |
| Score: | 0.55
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------GGATCACCAT
CGTTGGGGATTAGCCT |
|


|
|
PH0016.1_Cux1_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.55
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------GGATCACCAT-
ACCGGTTGATCACCTGA |
|


|
|
PB0153.1_Nr2f2_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.55
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------GGATCACCAT
CGCGCCGGGTCACGTA |
|


|
|
PH0035.1_Gsc/Jaspar
| Match Rank: | 10 |
| Score: | 0.55
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----GGATCACCAT--
NNAAGGGATTAACGANT |
|


|
|