| p-value: | 1e0 |
| log p-value: | -2.287e+00 |
| Information Content per bp: | 1.840 |
| Number of Target Sequences with motif | 6.0 |
| Percentage of Target Sequences with motif | 0.07% |
| Number of Background Sequences with motif | 3.1 |
| Percentage of Background Sequences with motif | 0.04% |
| Average Position of motif in Targets | 335.2 +/- 216.0bp |
| Average Position of motif in Background | 1310.4 +/- 57.5bp |
| Strand Bias (log2 ratio + to - strand density) | -1.0 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0143.3_Sox2/Jaspar
| Match Rank: | 1 |
| Score: | 0.60
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TCCTTAGTCG
-CCTTTGTT- |
|


|
|
MA0132.1_Pdx1/Jaspar
| Match Rank: | 2 |
| Score: | 0.57
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TCCTTAGTCG
-AATTAG--- |
|


|
|
Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer
| Match Rank: | 3 |
| Score: | 0.56
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TCCTTAGTCG
-CCWTTGTY- |
|


|
|
Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer
| Match Rank: | 4 |
| Score: | 0.55
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TCCTTAGTCG
BCMATTAG--- |
|


|
|
PAX3:FKHR-fusion(Paired/Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer
| Match Rank: | 5 |
| Score: | 0.55
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TCCTTAGTCG----
NNAATTAGTCACGGT |
|


|
|
PB0032.1_IRC900814_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.54
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TCCTTAGTCG-----
GNNATTTGTCGTAANN |
|


|
|
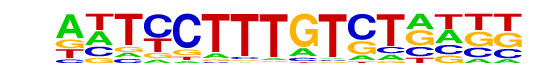
PB0166.1_Sox12_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.54
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TCCTTAGTCG----
ANTCCTTTGTCTNNNN |
|

|
|
MA0109.1_Hltf/Jaspar
| Match Rank: | 8 |
| Score: | 0.54
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TCCTTAGTCG
AACCTTATAT- |
|


|
|
PH0027.1_Emx2/Jaspar
| Match Rank: | 9 |
| Score: | 0.53
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----TCCTTAGTCG--
ACCACTAATTAGTGGAC |
|


|
|
PH0031.1_Evx1/Jaspar
| Match Rank: | 10 |
| Score: | 0.52
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----TCCTTAGTCG--
AGAACTAATTAGTGGAC |
|


|
|





















