| p-value: | 1e-10 |
| log p-value: | -2.475e+01 |
| Information Content per bp: | 1.857 |
| Number of Target Sequences with motif | 38.0 |
| Percentage of Target Sequences with motif | 0.54% |
| Number of Background Sequences with motif | 12.0 |
| Percentage of Background Sequences with motif | 0.15% |
| Average Position of motif in Targets | 335.3 +/- 301.9bp |
| Average Position of motif in Background | 671.7 +/- 510.2bp |
| Strand Bias (log2 ratio + to - strand density) | -0.4 |
| Multiplicity (# of sites on avg that occur together) | 1.03 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0164.1_Nr2e3/Jaspar
| Match Rank: | 1 |
| Score: | 0.67
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | TTCGAGCTTC
--CAAGCTT- |
|


|
|
POL008.1_DCE_S_I/Jaspar
| Match Rank: | 2 |
| Score: | 0.66
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | TTCGAGCTTC-
-----GCTTCC |
|


|
|
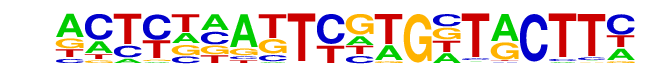
Mouse_Recombination_Hotspot/Testis-DMC1-ChIP-Seq(GSE24438)/Homer
| Match Rank: | 3 |
| Score: | 0.56
| | Offset: | -7
| | Orientation: | forward strand |
| Alignment: | -------TTCGAGCTTC---
ACTYKNATTCGTGNTACTTC |
|

|
|
MA0069.1_Pax6/Jaspar
| Match Rank: | 4 |
| Score: | 0.55
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TTCGAGCTTC----
TTCACGCATGAGTT |
|


|
|
PB0161.1_Rxra_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.55
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TTCGAGCTTC----
NNNNCAACCTTCGNGA |
|


|
|
MA0473.1_ELF1/Jaspar
| Match Rank: | 6 |
| Score: | 0.52
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TTCGAGCTTC
CACTTCCTGNTTC |
|


|
|
PB0090.1_Zbtb12_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.52
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------TTCGAGCTTC-
CTAAGGTTCTAGATCAC |
|


|
|
PB0014.1_Esrra_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.51
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TTCGAGCTTC----
NNNNATGACCTTGANTN |
|


|
|
Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer
| Match Rank: | 9 |
| Score: | 0.50
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TTCGAGCTTC
GKVTCADRTTWC |
|


|
|
MA0523.1_TCF7L2/Jaspar
| Match Rank: | 10 |
| Score: | 0.50
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTCGAGCTTC
TNCCTTTGATCTTN |
|


|
|





















