| p-value: | 1e-6 |
| log p-value: | -1.464e+01 |
| Information Content per bp: | 1.977 |
| Number of Target Sequences with motif | 20.0 |
| Percentage of Target Sequences with motif | 0.29% |
| Number of Background Sequences with motif | 6.6 |
| Percentage of Background Sequences with motif | 0.08% |
| Average Position of motif in Targets | 362.7 +/- 335.9bp |
| Average Position of motif in Background | 676.3 +/- 401.9bp |
| Strand Bias (log2 ratio + to - strand density) | 0.9 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0029.1_Hic1_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.77
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TGTTGGCATG--
NGTAGGTTGGCATNNN |
|


|
|
MA0161.1_NFIC/Jaspar
| Match Rank: | 2 |
| Score: | 0.72
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | TGTTGGCATG
--TTGGCA-- |
|


|
|
MA0600.1_RFX2/Jaspar
| Match Rank: | 3 |
| Score: | 0.68
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TGTTGGCATG-----
NNNCNGTTGCCATGGNAAC |
|


|
|
MA0510.1_RFX5/Jaspar
| Match Rank: | 4 |
| Score: | 0.68
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TGTTGGCATG---
NCTGTTGCCAGGGAG |
|


|
|
MA0509.1_Rfx1/Jaspar
| Match Rank: | 5 |
| Score: | 0.68
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TGTTGGCATG-----
-GTTGCCATGGNAAC |
|
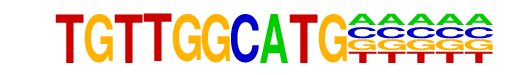

|
|
X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer
| Match Rank: | 6 |
| Score: | 0.68
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TGTTGGCATG----
GGTTGCCATGGCAA |
|

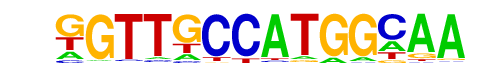
|
|
Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer
| Match Rank: | 7 |
| Score: | 0.67
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TGTTGGCATG----
NGTTGCCATGGCAA |
|


|
|
NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 8 |
| Score: | 0.67
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TGTTGGCATG
-CTTGGCAA- |
|


|
|
Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 9 |
| Score: | 0.66
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TGTTGGCATG-
CTGTTGCTAGGS |
|


|
|
Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer
| Match Rank: | 10 |
| Score: | 0.64
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TGTTGGCATG
GGTGYTGACAGS |
|


|
|





















