





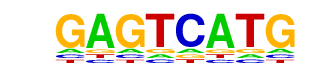


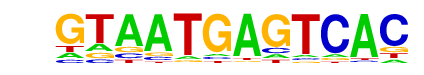

| p-value: | 1e-2 |
| log p-value: | -6.840e+00 |
| Information Content per bp: | 1.597 |
| Number of Target Sequences with motif | 143.0 |
| Percentage of Target Sequences with motif | 2.04% |
| Number of Background Sequences with motif | 129.5 |
| Percentage of Background Sequences with motif | 1.56% |
| Average Position of motif in Targets | 382.7 +/- 349.9bp |
| Average Position of motif in Background | 501.6 +/- 376.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.01 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.602 |  | 1e0 | -0.280652 | 7.26% | 7.48% | motif file (matrix) |
| 2 | 0.719 |  | 1e0 | -0.144052 | 3.10% | 3.33% | motif file (matrix) |
| 3 | 0.792 |  | 1e0 | -0.039059 | 0.87% | 1.09% | motif file (matrix) |
| 4 | 0.828 |  | 1e0 | -0.009491 | 4.82% | 5.45% | motif file (matrix) |
| 5 | 0.772 |  | 1e0 | -0.003667 | 4.29% | 4.97% | motif file (matrix) |
| 6 | 0.749 |  | 1e0 | -0.000353 | 3.48% | 4.28% | motif file (matrix) |
| 7 | 0.642 |  | 1e0 | -0.000000 | 4.31% | 5.66% | motif file (matrix) |
| 8 | 0.798 |  | 1e0 | -0.000000 | 2.44% | 3.76% | motif file (matrix) |
| 9 | 0.741 |  | 1e0 | -0.000000 | 20.18% | 23.64% | motif file (matrix) |