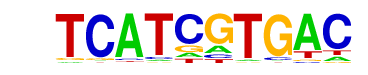
| p-value: | 1e-1 |
| log p-value: | -4.540e+00 |
| Information Content per bp: | 1.665 |
| Number of Target Sequences with motif | 180.0 |
| Percentage of Target Sequences with motif | 2.57% |
| Number of Background Sequences with motif | 178.3 |
| Percentage of Background Sequences with motif | 2.16% |
| Average Position of motif in Targets | 370.3 +/- 354.8bp |
| Average Position of motif in Background | 501.5 +/- 434.6bp |
| Strand Bias (log2 ratio + to - strand density) | -0.4 |
| Multiplicity (# of sites on avg that occur together) | 1.01 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
ERE(NR/IR3)/MCF7-ERa-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 1 |
| Score: | 0.73
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----TCATCGTGAC-
NAGGTCACNNTGACC |
|


|
|
MA0258.2_ESR2/Jaspar
| Match Rank: | 2 |
| Score: | 0.67
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TCATCGTGAC--
AGGNCANNGTGACCT |
|


|
|
MA0089.1_NFE2L1::MafG/Jaspar
| Match Rank: | 3 |
| Score: | 0.66
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TCATCGTGAC
GTCATN----- |
|


|
|
MA0112.2_ESR1/Jaspar
| Match Rank: | 4 |
| Score: | 0.66
| | Offset: | -8
| | Orientation: | forward strand |
| Alignment: | --------TCATCGTGAC--
GGCCCAGGTCACCCTGACCT |
|
|
|
bZIP:IRF/Th17-BatF-ChIP-Seq(GSE39756)/Homer
| Match Rank: | 5 |
| Score: | 0.63
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----TCATCGTGAC---
NAGTTTCABTHTGACTNW |
|


|
|
MA0078.1_Sox17/Jaspar
| Match Rank: | 6 |
| Score: | 0.61
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TCATCGTGAC
CTCATTGTC-- |
|


|
|
MA0067.1_Pax2/Jaspar
| Match Rank: | 7 |
| Score: | 0.61
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TCATCGTGAC-
---NCGTGACN |
|


|
|
MA0066.1_PPARG/Jaspar
| Match Rank: | 8 |
| Score: | 0.61
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----TCATCGTGAC-----
GTAGGTCACGGTGACCTACT |
|


|
|
PH0017.1_Cux1_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------TCATCGTGAC
TAGTGATCATCATTA- |
|


|
|
PAX5-shortForm(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 10 |
| Score: | 0.55
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TCATCGTGAC
TCAGNGAGCGTGAC |
|


|
|