| p-value: | 1e0 |
| log p-value: | -3.520e-02 |
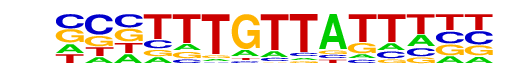
| Information Content per bp: | 1.703 |
| Number of Target Sequences with motif | 283.0 |
| Percentage of Target Sequences with motif | 4.04% |
| Number of Background Sequences with motif | 370.4 |
| Percentage of Background Sequences with motif | 4.48% |
| Average Position of motif in Targets | 450.4 +/- 434.2bp |
| Average Position of motif in Background | 504.9 +/- 406.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.07 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0182.1_Srf_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.80
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTTTTTTTCT---
NNNNTTTTTTTTTNAAC |
|


|
|
PB0192.1_Tcfap2e_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.79
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TTTTTTTTCT----
TTTTTTTTCNNGTN |
|


|
|
PB0093.1_Zfp105_1/Jaspar
| Match Rank: | 3 |
| Score: | 0.77
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTTTTTTTCT-
NTNTTGTTGTTTGTN |
|


|
|
PB0116.1_Elf3_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.74
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----TTTTTTTTCT--
GNATTTTTTTTTTGANC |
|


|
|
MA0465.1_CDX2/Jaspar
| Match Rank: | 5 |
| Score: | 0.71
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TTTTTTTTCT-
TTTTATGGCTN |
|


|
|
PH0078.1_Hoxd13/Jaspar
| Match Rank: | 6 |
| Score: | 0.70
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTTTTTTTCT--
NNANTTTTATTGGNNN |
|


|
|
PH0064.1_Hoxb9/Jaspar
| Match Rank: | 7 |
| Score: | 0.70
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTTTTTTTCT--
NGANTTTTATGGCTCN |
|


|
|
Cdx2(Homeobox)/mES-Cdx2-ChIP-Seq(GSE14586)/Homer
| Match Rank: | 8 |
| Score: | 0.70
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TTTTTTTTCT
NTTTTATGAC- |
|


|
|
PH0057.1_Hoxb13/Jaspar
| Match Rank: | 9 |
| Score: | 0.68
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTTTTTTTCT--
NNAATTTTATTGGNTN |
|


|
|
PB0119.1_Foxa2_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.68
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----TTTTTTTTCT
NCNTTTGTTATTTNN |
|

|
|





















