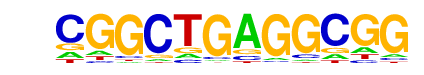
MA0162.2_EGR1/Jaspar
| Match Rank: | 1 |
| Score: | 0.67
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CGGCTGAGGCGG---
-GGCGGGGGCGGGGG |
|


|
|
MA0516.1_SP2/Jaspar
| Match Rank: | 2 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | reverse strand |
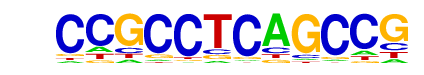
| Alignment: | CGGCTGAGGCGG---
GGGNGGGGGCGGGGC |
|


|
|
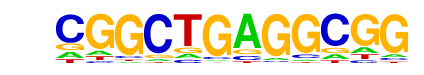
MA0117.1_Mafb/Jaspar
| Match Rank: | 3 |
| Score: | 0.65
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CGGCTGAGGCGG
--GCTGACGC-- |
|

|
|
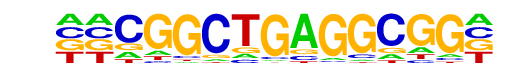
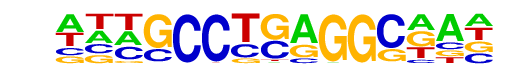
PB0088.1_Tcfap2e_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.63
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CGGCTGAGGCGG-
ATTGCCTGAGGCAAT |
|
|
|
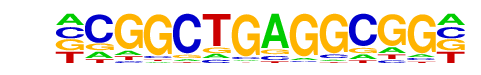
PB0010.1_Egr1_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.62
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CGGCTGAGGCGG-
ANTGCGGGGGCGGN |
|

|
|
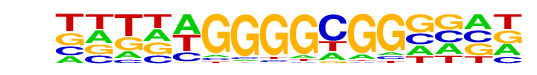
PB0110.1_Bcl6b_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CGGCTGAGGCGG----
NNTNAGGGGCGGNNNN |
|

|
|
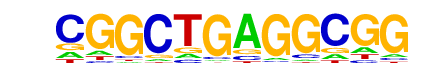
Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 7 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CGGCTGAGGCGG
TGGGTGTGGC-- |
|

|
|
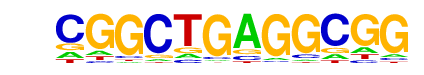
MA0039.2_Klf4/Jaspar
| Match Rank: | 8 |
| Score: | 0.58
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGGCTGAGGCGG
TGGGTGGGGC-- |
|

|
|
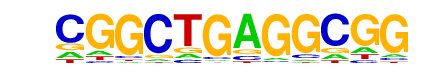
KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer
| Match Rank: | 9 |
| Score: | 0.58
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGGCTGAGGCGG
DGGGYGKGGC-- |
|

|
|
MA0079.3_SP1/Jaspar
| Match Rank: | 10 |
| Score: | 0.58
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CGGCTGAGGCGG---
----GGGGGCGGGGC |
|


|
|