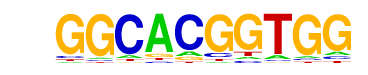
| p-value: | 1e-7 |
| log p-value: | -1.686e+01 |
| Information Content per bp: | 1.649 |
| Number of Target Sequences with motif | 830.0 |
| Percentage of Target Sequences with motif | 18.53% |
| Number of Background Sequences with motif | 3894.1 |
| Percentage of Background Sequences with motif | 15.56% |
| Average Position of motif in Targets | 565.5 +/- 501.6bp |
| Average Position of motif in Background | 566.6 +/- 509.3bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.31 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
POL006.1_BREu/Jaspar
| Match Rank: | 1 |
| Score: | 0.65
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CCACCGTGCC
--AGCGCGCC |
|


|
|
MA0088.1_znf143/Jaspar
| Match Rank: | 2 |
| Score: | 0.63
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------CCACCGTGCC----
GATTTCCCATAATGCCTTGC |
|


|
|
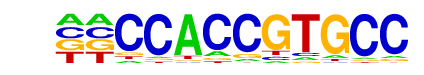
MA0130.1_ZNF354C/Jaspar
| Match Rank: | 3 |
| Score: | 0.59
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CCACCGTGCC
ATCCAC------ |
|

|
|
MA0119.1_TLX1::NFIC/Jaspar
| Match Rank: | 4 |
| Score: | 0.58
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CCACCGTGCC--
TGGCACCATGCCAA |
|


|
|
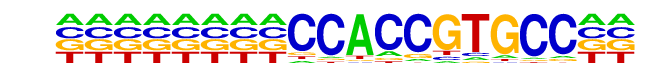
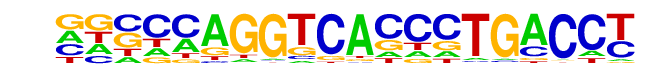
MA0112.2_ESR1/Jaspar
| Match Rank: | 5 |
| Score: | 0.56
| | Offset: | -8
| | Orientation: | forward strand |
| Alignment: | --------CCACCGTGCC--
GGCCCAGGTCACCCTGACCT |
|
|
|
ERE(NR/IR3)/MCF7-ERa-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 6 |
| Score: | 0.55
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CCACCGTGCC-
NAGGTCACNNTGACC |
|


|
|
MA0259.1_HIF1A::ARNT/Jaspar
| Match Rank: | 7 |
| Score: | 0.55
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CCACCGTGCC
-GGACGTGC- |
|


|
|
ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer
| Match Rank: | 8 |
| Score: | 0.55
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CCACCGTGCC
ATTTCCCAGVAKSCY |
|


|
|
SD0002.1_at_AC_acceptor/Jaspar
| Match Rank: | 9 |
| Score: | 0.54
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CCACCGTGCC--
-NNACTTGCCTT |
|


|
|
PB0196.1_Zbtb7b_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.54
| | Offset: | -7
| | Orientation: | forward strand |
| Alignment: | -------CCACCGTGCC
CATAAGACCACCATTAC |
|


|
|