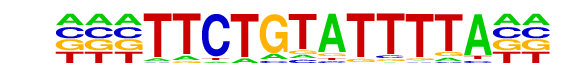
PB0106.1_Arid5a_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.61
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TTCTGTATTTTA--
TNNTTTCGTATTNNANN |
|

|
|
PH0046.1_Hoxa10/Jaspar
| Match Rank: | 2 |
| Score: | 0.57
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TTCTGTATTTTA-------
---TNAATTTTATTACCTN |
|


|
|
PH0078.1_Hoxd13/Jaspar
| Match Rank: | 3 |
| Score: | 0.56
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TTCTGTATTTTA-------
---NNANTTTTATTGGNNN |
|


|
|
PH0075.1_Hoxd10/Jaspar
| Match Rank: | 4 |
| Score: | 0.56
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TTCTGTATTTTA-------
--NTNAATTTTATTGNATT |
|


|
|
MA0479.1_FOXH1/Jaspar
| Match Rank: | 5 |
| Score: | 0.56
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TTCTGTATTTTA
-TGTGGATTNNN |
|


|
|
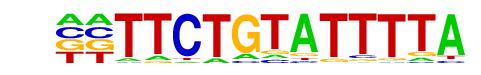
PB0162.1_Sfpi1_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.55
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TTCTGTATTTTA
GGTTCCNNAATTTG |
|

|
|
HOXD13(Homeobox)/Chicken-Hoxd13-ChIP-Seq(GSE38910)/Homer
| Match Rank: | 7 |
| Score: | 0.55
| | Offset: | 7
| | Orientation: | reverse strand |
| Alignment: | TTCTGTATTTTA-----
-------TTTTATTRGN |
|


|
|
Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 8 |
| Score: | 0.55
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TTCTGTATTTTA---
---KCTATTTTTRGH |
|


|
|
PH0064.1_Hoxb9/Jaspar
| Match Rank: | 9 |
| Score: | 0.54
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TTCTGTATTTTA-------
---NGANTTTTATGGCTCN |
|


|
|
POL007.1_BREd/Jaspar
| Match Rank: | 10 |
| Score: | 0.54
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | TTCTGTATTTTA
----GTTTGTT- |
|


|
|





















