MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer
| Match Rank: | 1 |
| Score: | 0.61
| | Offset: | 3
| | Orientation: | forward strand |
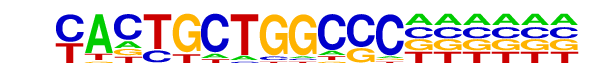
| Alignment: | YACTGCTGGCCC-
---TGCTGACTCA |
|


|
|
MA0461.1_Atoh1/Jaspar
| Match Rank: | 2 |
| Score: | 0.58
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | YACTGCTGGCCC
--CAGATGGC-- |
|


|
|
MA0161.1_NFIC/Jaspar
| Match Rank: | 3 |
| Score: | 0.56
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | YACTGCTGGCCC
-----TTGGCA- |
|


|
|
POL010.1_DCE_S_III/Jaspar
| Match Rank: | 4 |
| Score: | 0.55
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | YACTGCTGGCCC
---NGCTN---- |
|


|
|
MA0496.1_MAFK/Jaspar
| Match Rank: | 5 |
| Score: | 0.54
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -YACTGCTGGCCC--
AAANTGCTGACTNAG |
|


|
|
MA0495.1_MAFF/Jaspar
| Match Rank: | 6 |
| Score: | 0.53
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---YACTGCTGGCCC---
NAAAANTGCTGACTCAGC |
|


|
|
PB0196.1_Zbtb7b_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.53
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -YACTGCTGGCCC----
NNANTGGTGGTCTTNNN |
|


|
|
PB0112.1_E2F2_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.53
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | YACTGCTGGCCC------
-NNNNTTGGCGCCGANNN |
|

|
|
MafF(bZIP)/HepG2-MafF-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 9 |
| Score: | 0.52
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --YACTGCTGGCCC-
AAAWWTGCTGACWWD |
|


|
|
MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer
| Match Rank: | 10 |
| Score: | 0.52
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | YACTGCTGGCCC
AACAGCTG---- |
|


|
|





















