| p-value: | 1e-11 |
| log p-value: | -2.533e+01 |
| Information Content per bp: | 1.707 |
| Number of Target Sequences with motif | 637.0 |
| Percentage of Target Sequences with motif | 14.22% |
| Number of Background Sequences with motif | 2742.8 |
| Percentage of Background Sequences with motif | 10.96% |
| Average Position of motif in Targets | 485.4 +/- 472.1bp |
| Average Position of motif in Background | 481.1 +/- 465.8bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.16 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0208.1_Zscan4_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.71
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GCACAHCAGC--
CGAAGCACACAAAATA |
|


|
|
PB0130.1_Gm397_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.64
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GCACAHCAGC--
AGCGGCACACACGCAA |
|


|
|
PB0120.1_Foxj1_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.63
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GCACAHCAGC--
ATGTCACAACAACAC |
|


|
|
PB0207.1_Zic3_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.61
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GCACAHCAGC---
GAGCACAGCAGGACA |
|


|
|
Nkx2.1(Homeobox)/LungAC-Nkx2.1-ChIP-Seq(GSE43252)/Homer
| Match Rank: | 5 |
| Score: | 0.61
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GCACAHCAGC
RSCACTYRAG- |
|


|
|
PB0044.1_Mtf1_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.60
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----GCACAHCAGC-
NNTTTGCACACGGCCC |
|


|
|
Unknown-ESC-element/mES-Nanog-ChIP-Seq(GSE11724)/Homer
| Match Rank: | 7 |
| Score: | 0.60
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GCACAHCAGC---
-CACAGCAGGGGG |
|


|
|
PB0205.1_Zic1_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.59
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GCACAHCAGC---
CCACACAGCAGGAGA |
|


|
|
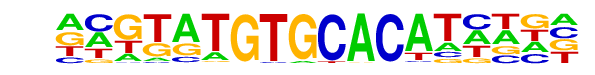
PB0026.1_Gm397_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.59
| | Offset: | -8
| | Orientation: | reverse strand |
| Alignment: | --------GCACAHCAGC
NNGTATGTGCACATNNN- |
|

|
|
MA0596.1_SREBF2/Jaspar
| Match Rank: | 10 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GCACAHCAGC
ATCACCCCAT- |
|


|
|





















