ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer
| Match Rank: | 1 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ACTACAACCT--
ACCACATCCTGT |
|


|
|
PB0090.1_Zbtb12_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.62
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----ACTACAACCT---
NNGATCTAGAACCTNNN |
|


|
|
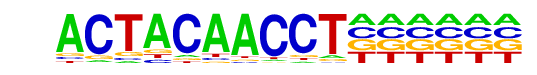
PB0161.1_Rxra_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ACTACAACCT------
NNNNCAACCTTCGNGA |
|

|
|
PB0122.1_Foxk1_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.59
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ACTACAACCT--
CAAACAACAACACCT |
|


|
|
PB0181.1_Spdef_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.59
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -ACTACAACCT-----
GATAACATCCTAGTAG |
|


|
|
MA0133.1_BRCA1/Jaspar
| Match Rank: | 6 |
| Score: | 0.59
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | ACTACAACCT
---ACAACAC |
|


|
|
MA0027.1_En1/Jaspar
| Match Rank: | 7 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ACTACAACCT
GANCACTACTT |
|


|
|
MA0493.1_Klf1/Jaspar
| Match Rank: | 8 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -ACTACAACCT
GGCCACACCCA |
|


|
|
RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer
| Match Rank: | 9 |
| Score: | 0.57
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ACTACAACCT
NWAACCACADNN- |
|


|
|
EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer
| Match Rank: | 10 |
| Score: | 0.57
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ACTACAACCT-
GGCCACACCCAN |
|


|
|





















