PB0010.1_Egr1_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.74
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GCTGAGGCGG-
ANTGCGGGGGCGGN |
|


|
|
MA0162.2_EGR1/Jaspar
| Match Rank: | 2 |
| Score: | 0.73
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GCTGAGGCGG---
GGCGGGGGCGGGGG |
|


|
|
MA0117.1_Mafb/Jaspar
| Match Rank: | 3 |
| Score: | 0.71
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GCTGAGGCGG
GCTGACGC-- |
|


|
|
MA0516.1_SP2/Jaspar
| Match Rank: | 4 |
| Score: | 0.68
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GCTGAGGCGG---
GGGNGGGGGCGGGGC |
|


|
|
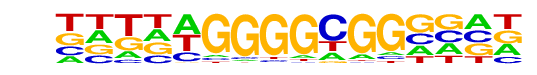
PB0110.1_Bcl6b_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.66
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GCTGAGGCGG----
NNTNAGGGGCGGNNNN |
|

|
|
MA0079.3_SP1/Jaspar
| Match Rank: | 6 |
| Score: | 0.64
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GCTGAGGCGG---
--GGGGGCGGGGC |
|


|
|
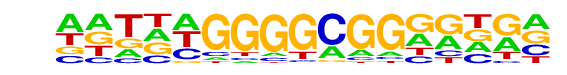
PB0202.1_Zfp410_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.62
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GCTGAGGCGG-----
NNTNNGGGGCGGNGNGN |
|

|
|
MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer
| Match Rank: | 8 |
| Score: | 0.60
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GCTGAGGCGG
TGCTGACTCA- |
|


|
|
MA0495.1_MAFF/Jaspar
| Match Rank: | 9 |
| Score: | 0.60
| | Offset: | -7
| | Orientation: | reverse strand |
| Alignment: | -------GCTGAGGCGG-
NAAAANTGCTGACTCAGC |
|


|
|
Sp1(Zf)/Promoter/Homer
| Match Rank: | 10 |
| Score: | 0.60
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GCTGAGGCGG----
--GGGGGCGGGGCC |
|


|
|





















