

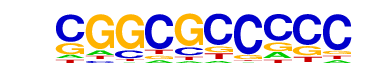



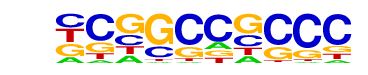











| p-value: | 1e-2 |
| log p-value: | -6.279e+00 |
| Information Content per bp: | 1.635 |
| Number of Target Sequences with motif | 68.0 |
| Percentage of Target Sequences with motif | 1.52% |
| Number of Background Sequences with motif | 82.1 |
| Percentage of Background Sequences with motif | 1.04% |
| Average Position of motif in Targets | 858.7 +/- 578.9bp |
| Average Position of motif in Background | 812.4 +/- 446.0bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.18 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.753 |  | 1e-2 | -5.760805 | 0.58% | 0.32% | motif file (matrix) |
| 2 | 0.614 |  | 1e-2 | -5.264631 | 6.19% | 5.31% | motif file (matrix) |
| 3 | 0.650 |  | 1e-2 | -5.139969 | 0.69% | 0.43% | motif file (matrix) |
| 4 | 0.864 |  | 1e-2 | -4.726022 | 0.18% | 0.07% | motif file (matrix) |
| 5 | 0.668 |  | 1e-1 | -2.717559 | 3.58% | 3.18% | motif file (matrix) |
| 6 | 0.784 |  | 1e-1 | -2.354439 | 1.41% | 1.20% | motif file (matrix) |
| 7 | 0.675 |  | 1e0 | -2.079125 | 0.94% | 0.78% | motif file (matrix) |
| 8 | 0.868 |  | 1e0 | -1.825767 | 0.47% | 0.38% | motif file (matrix) |
| 9 | 0.698 |  | 1e0 | -1.690418 | 0.56% | 0.46% | motif file (matrix) |
| 10 | 0.688 |  | 1e0 | -1.411405 | 0.36% | 0.30% | motif file (matrix) |
| 11 | 0.616 |  | 1e0 | -1.328673 | 2.88% | 2.72% | motif file (matrix) |
| 12 | 0.685 |  | 1e0 | -0.906196 | 2.66% | 2.61% | motif file (matrix) |
| 13 | 0.675 |  | 1e0 | -0.684702 | 0.69% | 0.69% | motif file (matrix) |
| 14 | 0.633 |  | 1e0 | -0.180204 | 1.43% | 1.61% | motif file (matrix) |
| 15 | 0.637 |  | 1e0 | -0.167284 | 7.85% | 8.26% | motif file (matrix) |
| 16 | 0.658 |  | 1e0 | -0.154931 | 0.92% | 1.08% | motif file (matrix) |