PB0039.1_Klf7_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.62
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CGGGCATGGTGG-
NNAGGGGCGGGGTNNA |
|


|
|
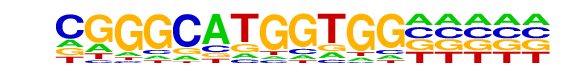
ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer
| Match Rank: | 2 |
| Score: | 0.61
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CGGGCATGGTGG-----
--RGSMTBCTGGGAAAT |
|

|
|
MA0088.1_znf143/Jaspar
| Match Rank: | 3 |
| Score: | 0.61
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CGGGCATGGTGG------
GCAAGGCATGATGGGAAATC |
|


|
|
GFY(?)/Promoter/Homer
| Match Rank: | 4 |
| Score: | 0.58
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CGGGCATGGTGG-
-GGGAATTGTAGT |
|


|
|
MA0597.1_THAP1/Jaspar
| Match Rank: | 5 |
| Score: | 0.58
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CGGGCATGGTGG
TNNGGGCAG----- |
|


|
|
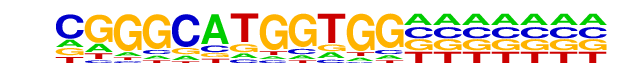
PB0196.1_Zbtb7b_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.58
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CGGGCATGGTGG-------
--NNANTGGTGGTCTTNNN |
|

|
|
MA0130.1_ZNF354C/Jaspar
| Match Rank: | 7 |
| Score: | 0.57
| | Offset: | 8
| | Orientation: | reverse strand |
| Alignment: | CGGGCATGGTGG--
--------GTGGAT |
|


|
|
POL006.1_BREu/Jaspar
| Match Rank: | 8 |
| Score: | 0.56
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CGGGCATGGTGG
--GGCGCGCT-- |
|


|
|
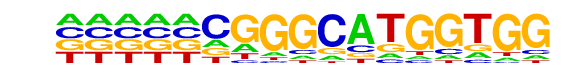
p53(p53)/mES-cMyc-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----CGGGCATGGTGG
ATGCCCGGGCATGT--- |
|

|
|
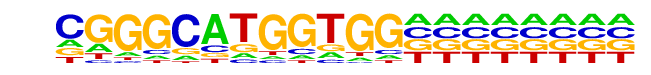
MA0112.2_ESR1/Jaspar
| Match Rank: | 10 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CGGGCATGGTGG--------
AGGTCAGGGTGACCTGGNNN |
|

|
|





















