PB0008.1_E2F2_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.72
| | Offset: | -7
| | Orientation: | forward strand |
| Alignment: | -------CGCGCGAC
ATAAAGGCGCGCGAT |
|


|
|
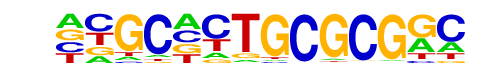
PB0199.1_Zfp161_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.71
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------CGCGCGAC
NNGCNCTGCGCGGC |
|

|
|
PB0095.1_Zfp161_1/Jaspar
| Match Rank: | 3 |
| Score: | 0.71
| | Offset: | -7
| | Orientation: | reverse strand |
| Alignment: | -------CGCGCGAC-
NCANGCGCGCGCGCCA |
|


|
|
PB0009.1_E2F3_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.71
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CGCGCGAC-----
ANCGCGCGCCCTTNN |
|


|
|
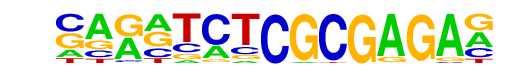
MA0527.1_ZBTB33/Jaspar
| Match Rank: | 5 |
| Score: | 0.69
| | Offset: | -7
| | Orientation: | reverse strand |
| Alignment: | -------CGCGCGAC
NAGNTCTCGCGAGAN |
|

|
|
E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 6 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGCGCGAC--
GGCGGGAAAH |
|


|
|
ZBTB33(Zf)/GM12878-ZBTB33-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------CGCGCGAC-
GGNTCTCGCGAGAAC |
|


|
|
GFX(?)/Promoter/Homer
| Match Rank: | 8 |
| Score: | 0.64
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CGCGCGAC
ATTCTCGCGAGA- |
|


|
|
MA0024.2_E2F1/Jaspar
| Match Rank: | 9 |
| Score: | 0.64
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CGCGCGAC-
CGGGCGGGAGG |
|


|
|
E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer
| Match Rank: | 10 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CGCGCGAC---
TGGCGGGAAAHB |
|


|
|





















