| p-value: | 1e0 |
| log p-value: | -4.661e-01 |
| Information Content per bp: | 1.910 |
| Number of Target Sequences with motif | 90.0 |
| Percentage of Target Sequences with motif | 2.01% |
| Number of Background Sequences with motif | 163.7 |
| Percentage of Background Sequences with motif | 2.08% |
| Average Position of motif in Targets | 354.8 +/- 375.2bp |
| Average Position of motif in Background | 536.0 +/- 479.4bp |
| Strand Bias (log2 ratio + to - strand density) | 0.3 |
| Multiplicity (# of sites on avg that occur together) | 1.03 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 1 |
| Score: | 0.65
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GATCATGC-
ACATCCTGNT |
|


|
|
MA0089.1_NFE2L1::MafG/Jaspar
| Match Rank: | 2 |
| Score: | 0.59
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GATCATGC
-GTCATN- |
|


|
|
PH0016.1_Cux1_1/Jaspar
| Match Rank: | 3 |
| Score: | 0.57
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------GATCATGC---
TNAGNTGATCAACCGGT |
|


|
|
PH0017.1_Cux1_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.56
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----GATCATGC---
TAGTGATCATCATTA |
|


|
|
PB0207.1_Zic3_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.55
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GATCATGC-------
NNTCCTGCTGTGNNN |
|


|
|
HNF6(Homeobox)/Liver-Hnf6-ChIP-Seq(ERP000394)/Homer
| Match Rank: | 6 |
| Score: | 0.55
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----GATCATGC
NTATYGATCH--- |
|


|
|
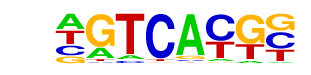
MA0067.1_Pax2/Jaspar
| Match Rank: | 7 |
| Score: | 0.54
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GATCATGC
AGTCACGC |
|

|
|
NRF1/Promoter/Homer
| Match Rank: | 8 |
| Score: | 0.54
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GATCATGC----
GCGCATGCGCAC |
|


|
|
PRDM9(Zf)/Testis-DMC1-ChIP-Seq(GSE35498)/Homer
| Match Rank: | 9 |
| Score: | 0.53
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GATCATGC------
AGATGCTRCTRCCHT |
|


|
|
MA0506.1_NRF1/Jaspar
| Match Rank: | 10 |
| Score: | 0.52
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GATCATGC---
GCGCCTGCGCA |
|


|
|





















