| p-value: | 1e-5 |
| log p-value: | -1.156e+01 |
| Information Content per bp: | 1.698 |
| Number of Target Sequences with motif | 453.0 |
| Percentage of Target Sequences with motif | 8.37% |
| Number of Background Sequences with motif | 1610.8 |
| Percentage of Background Sequences with motif | 6.85% |
| Average Position of motif in Targets | 419.3 +/- 402.2bp |
| Average Position of motif in Background | 448.8 +/- 413.6bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.08 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer
| Match Rank: | 1 |
| Score: | 0.58
| | Offset: | 1
| | Orientation: | forward strand |
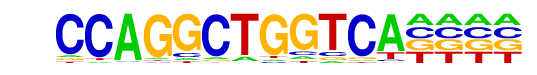
| Alignment: | CCAGGCTGGTCA
-TWGTCTGV--- |
|


|
|
GLI3(Zf)/GLI3-ChIP-Chip(GSE11077)/Homer
| Match Rank: | 2 |
| Score: | 0.58
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CCAGGCTGGTCA
CGTGGGTGGTCC |
|


|
|
Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer
| Match Rank: | 3 |
| Score: | 0.57
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CCAGGCTGGTCA
CCAGACAG---- |
|


|
|
PB0153.1_Nr2f2_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.55
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CCAGGCTGGTCA----
CGCGCCGGGTCACGTA |
|

|
|
SD0002.1_at_AC_acceptor/Jaspar
| Match Rank: | 5 |
| Score: | 0.54
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CCAGGCTGGTCA
-AAGGCAAGTGT |
|


|
|
PB0060.1_Smad3_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.54
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CCAGGCTGGTCA
CAAATCCAGACATCACA |
|


|
|
MF0004.1_Nuclear_Receptor_class/Jaspar
| Match Rank: | 7 |
| Score: | 0.54
| | Offset: | 6
| | Orientation: | forward strand |
| Alignment: | CCAGGCTGGTCA
------AGGTCA |
|


|
|
POL010.1_DCE_S_III/Jaspar
| Match Rank: | 8 |
| Score: | 0.54
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CCAGGCTGGTCA
---NGCTN---- |
|


|
|
Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer
| Match Rank: | 9 |
| Score: | 0.53
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CCAGGCTGGTCA
CCAGACRSVB-- |
|


|
|
PB0196.1_Zbtb7b_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.53
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCAGGCTGGTCA----
NNANTGGTGGTCTTNNN |
|


|
|





















