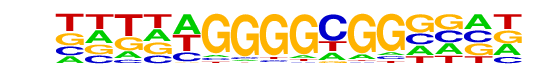
PB0092.1_Zbtb7b_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.64
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTGGGAGGCT-
ATTTTNGGGGGGCNN |
|


|
|
MA0503.1_Nkx2-5_(var.2)/Jaspar
| Match Rank: | 2 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TTGGGAGGCT
CTTGAGTGGCT |
|


|
|
PB0167.1_Sox13_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.61
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TTGGGAGGCT----
GTATTGGGTGGGTAATT |
|


|
|
PB0201.1_Zfp281_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.59
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTGGGAGGCT---
NNNATTGGGGGTNTCCT |
|


|
|
MA0161.1_NFIC/Jaspar
| Match Rank: | 5 |
| Score: | 0.57
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TTGGGAGGCT
TTGGCA---- |
|


|
|
POL010.1_DCE_S_III/Jaspar
| Match Rank: | 6 |
| Score: | 0.57
| | Offset: | 6
| | Orientation: | reverse strand |
| Alignment: | TTGGGAGGCT-
------NGCTN |
|


|
|
PB0203.1_Zfp691_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.56
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TTGGGAGGCT----
NTNNNAGGAGTCTCNTN |
|


|
|
Egr2/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer
| Match Rank: | 8 |
| Score: | 0.55
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TTGGGAGGCT
NGCGTGGGCGGR- |
|


|
|
NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 9 |
| Score: | 0.54
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TTGGGAGGCT
CTTGGCAA--- |
|


|
|
PB0110.1_Bcl6b_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.54
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTGGGAGGCT--
NNTNAGGGGCGGNNNN |
|

|
|





















