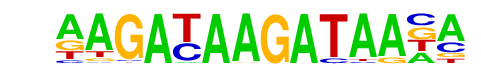MA0037.2_GATA3/Jaspar
| Match Rank: | 1 |
| Score: | 0.65
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AAGAAAAGAACA
-AGATAAGA--- |
|


|
|
MA0029.1_Mecom/Jaspar
| Match Rank: | 2 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAGAAAAGAACA--
AAGATAAGATAACA |
|

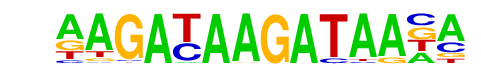
|
|
MA0035.3_Gata1/Jaspar
| Match Rank: | 3 |
| Score: | 0.64
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AAGAAAAGAACA
ANAGATAAGAA-- |
|


|
|
MA0036.2_GATA2/Jaspar
| Match Rank: | 4 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AAGAAAAGAACA-
NCAGATAAGAANNN |
|


|
|
MA0041.1_Foxd3/Jaspar
| Match Rank: | 5 |
| Score: | 0.63
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | AAGAAAAGAACA---
---AAACAAACATTC |
|


|
|
PB0093.1_Zfp105_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.62
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AAGAAAAGAACA----
-AACAAACAACAAGAG |
|


|
|
PB0119.1_Foxa2_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.62
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | AAGAAAAGAACA-----
--AAAAATAACAAACGG |
|


|
|
PB0021.1_Gata3_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.61
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------AAGAAAAGAACA----
TTTTTAGAGATAAGAAATAAAG |
|


|
|
MA0482.1_Gata4/Jaspar
| Match Rank: | 9 |
| Score: | 0.61
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AAGAAAAGAACA
NNGAGATAAGA--- |
|


|
|
PB0122.1_Foxk1_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAGAAAAGAACA---
CAAACAACAACACCT |
|


|
|