PB0008.1_E2F2_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.82
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TAGCGCGC------
NTCGCGCGCCTTNNN |
|


|
|
PB0009.1_E2F3_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.77
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TAGCGCGC------
ANCGCGCGCCCTTNN |
|


|
|
POL006.1_BREu/Jaspar
| Match Rank: | 3 |
| Score: | 0.74
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TAGCGCGC-
-AGCGCGCC |
|


|
|
PB0095.1_Zfp161_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.70
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TAGCGCGC------
TGGCGCGCGCGCCTGA |
|


|
|
MA0024.2_E2F1/Jaspar
| Match Rank: | 5 |
| Score: | 0.64
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TAGCGCGC-
CCTCCCGCCCN |
|


|
|
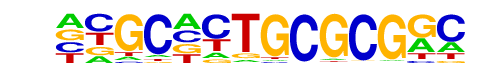
PB0199.1_Zfp161_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.62
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----TAGCGCGC-
NNGCNCTGCGCGGC |
|

|
|
Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo et al.)/Homer
| Match Rank: | 7 |
| Score: | 0.61
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TAGCGCGC--
TTGCGTGCVA |
|


|
|
PB0112.1_E2F2_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.59
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----TAGCGCGC----
NNNNTTGGCGCCGANNN |
|


|
|
PB0113.1_E2F3_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.59
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----TAGCGCGC----
NNNNTTGGCGCCGANNN |
|


|
|
E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 10 |
| Score: | 0.59
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TAGCGCGC
DTTTCCCGCC- |
|


|
|





















