


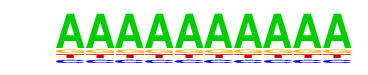






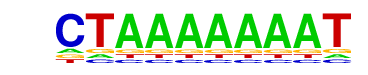

| p-value: | 1e-7 |
| log p-value: | -1.707e+01 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 40.0 |
| Percentage of Target Sequences with motif | 0.74% |
| Number of Background Sequences with motif | 64.3 |
| Percentage of Background Sequences with motif | 0.27% |
| Average Position of motif in Targets | 346.4 +/- 341.0bp |
| Average Position of motif in Background | 550.5 +/- 489.3bp |
| Strand Bias (log2 ratio + to - strand density) | 0.6 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.674 |  | 1e-1 | -4.023187 | 3.09% | 2.62% | motif file (matrix) |
| 2 | 0.789 |  | 1e-1 | -2.897369 | 1.57% | 1.31% | motif file (matrix) |
| 3 | 0.609 |  | 1e0 | -0.639335 | 18.28% | 18.31% | motif file (matrix) |
| 4 | 0.667 |  | 1e0 | -0.250229 | 14.30% | 14.66% | motif file (matrix) |
| 5 | 0.636 |  | 1e0 | -0.224039 | 4.07% | 4.29% | motif file (matrix) |
| 6 | 0.745 |  | 1e0 | -0.165026 | 21.64% | 22.21% | motif file (matrix) |
| 7 | 0.838 |  | 1e0 | -0.113853 | 15.34% | 15.95% | motif file (matrix) |
| 8 | 0.680 |  | 1e0 | -0.091547 | 3.31% | 3.64% | motif file (matrix) |
| 9 | 0.793 |  | 1e0 | -0.040759 | 10.66% | 11.41% | motif file (matrix) |
| 10 | 0.684 |  | 1e0 | -0.015098 | 9.07% | 9.94% | motif file (matrix) |