




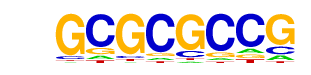


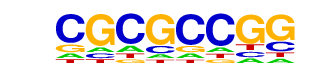


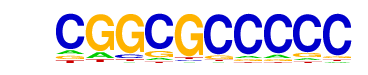

| p-value: | 1e-2 |
| log p-value: | -5.219e+00 |
| Information Content per bp: | 1.651 |
| Number of Target Sequences with motif | 25.0 |
| Percentage of Target Sequences with motif | 0.46% |
| Number of Background Sequences with motif | 21.5 |
| Percentage of Background Sequences with motif | 0.27% |
| Average Position of motif in Targets | 824.0 +/- 509.4bp |
| Average Position of motif in Background | 783.1 +/- 375.1bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.05 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.638 |  | 1e-1 | -4.239256 | 2.89% | 2.42% | motif file (matrix) |
| 2 | 0.743 |  | 1e-1 | -3.499576 | 0.50% | 0.34% | motif file (matrix) |
| 3 | 0.862 |  | 1e0 | -1.979962 | 4.40% | 4.11% | motif file (matrix) |
| 4 | 0.703 |  | 1e0 | -0.900838 | 4.07% | 4.01% | motif file (matrix) |
| 5 | 0.798 |  | 1e0 | -0.754209 | 2.22% | 2.20% | motif file (matrix) |
| 6 | 0.755 |  | 1e0 | -0.641735 | 1.50% | 1.51% | motif file (matrix) |
| 7 | 0.760 |  | 1e0 | -0.626417 | 2.53% | 2.55% | motif file (matrix) |
| 8 | 0.720 |  | 1e0 | -0.268345 | 1.33% | 1.44% | motif file (matrix) |
| 9 | 0.652 |  | 1e0 | -0.110400 | 1.39% | 1.59% | motif file (matrix) |
| 10 | 0.671 |  | 1e0 | -0.032371 | 1.72% | 2.06% | motif file (matrix) |
| 11 | 0.638 |  | 1e0 | -0.012375 | 1.79% | 2.22% | motif file (matrix) |