| p-value: | 1e-1 |
| log p-value: | -4.032e+00 |
| Information Content per bp: | 1.868 |
| Number of Target Sequences with motif | 560.0 |
| Percentage of Target Sequences with motif | 10.36% |
| Number of Background Sequences with motif | 765.7 |
| Percentage of Background Sequences with motif | 9.51% |
| Average Position of motif in Targets | 423.0 +/- 418.1bp |
| Average Position of motif in Background | 548.0 +/- 471.6bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.27 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Pitx1(Homeobox)/Chicken-Pitx1-ChIP-Seq(GSE38910)/Homer
| Match Rank: | 1 |
| Score: | 0.76
| | Offset: | 2
| | Orientation: | reverse strand |
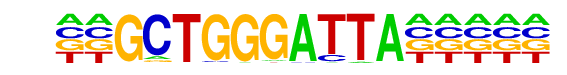
| Alignment: | GCTGGGATTA
--NGGGATTA |
|


|
|
CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer
| Match Rank: | 2 |
| Score: | 0.73
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | GCTGGGATTA--
----GGATTAGC |
|


|
|
PB0185.1_Tcf1_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.73
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GCTGGGATTA--
TTGCCCGGATTAGG |
|


|
|
PH0129.1_Otx1/Jaspar
| Match Rank: | 4 |
| Score: | 0.72
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GCTGGGATTA------
AGAGGGGATTAATTTAT |
|


|
|
PH0137.1_Pitx1/Jaspar
| Match Rank: | 5 |
| Score: | 0.72
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GCTGGGATTA-----
TTAGAGGGATTAACAAT |
|

|
|
PH0138.1_Pitx2/Jaspar
| Match Rank: | 6 |
| Score: | 0.72
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GCTGGGATTA------
TGAAGGGATTAATCATC |
|


|
|
PH0124.1_Obox5_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.72
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GCTGGGATTA------
TAGAGGGATTAAATTTC |
|


|
|
PH0130.1_Otx2/Jaspar
| Match Rank: | 8 |
| Score: | 0.72
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GCTGGGATTA------
TGTAGGGATTAATTGTC |
|


|
|
MA0483.1_Gfi1b/Jaspar
| Match Rank: | 9 |
| Score: | 0.71
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GCTGGGATTA
TGCTGTGATTT |
|


|
|
Gfi1b(Zf)/HPC7-Gfi1b-ChIP-Seq(GSE22178)/Homer
| Match Rank: | 10 |
| Score: | 0.71
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GCTGGGATTA
GCAGTGATTT |
|


|
|





















