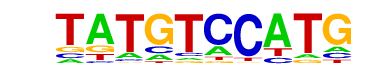
Nr5a2(NR)/Pancreas-LRH1-ChIP-Seq(GSE34295)/Homer
| Match Rank: | 1 |
| Score: | 0.65
| | Offset: | 2
| | Orientation: | reverse strand |
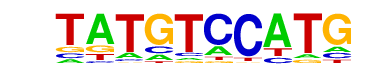
| Alignment: | TATGTCCATG--
--TGACCTTGAV |
|

|
|
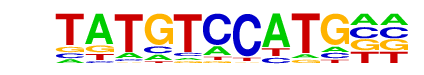
Nr5a2(NR)/mES-Nr5a2-ChIP-Seq(GSE19019)/Homer
| Match Rank: | 2 |
| Score: | 0.64
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TATGTCCATG--
--TGACCTTGAN |
|

|
|
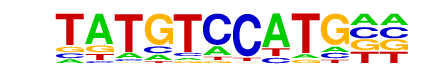
SD0003.1_at_AC_acceptor/Jaspar
| Match Rank: | 3 |
| Score: | 0.64
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TATGTCCATG
AAGATATCCTT- |
|

|
|
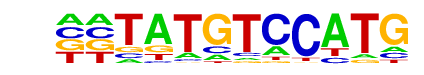
MA0507.1_POU2F2/Jaspar
| Match Rank: | 4 |
| Score: | 0.63
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TATGTCCATG-
TTCATTTGCATAT |
|

|
|
AR-halfsite(NR)/LNCaP-AR-ChIP-Seq(GSE27824)/Homer
| Match Rank: | 5 |
| Score: | 0.63
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TATGTCCATG-
-CTGTTCCTGG |
|

|
|
PB0014.1_Esrra_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.63
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TATGTCCATG----
NNNNATGACCTTGANTN |
|

|
|
MA0592.1_ESRRA/Jaspar
| Match Rank: | 7 |
| Score: | 0.62
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TATGTCCATG-
NGTGACCTTGG |
|

|
|
PH0082.1_Irx2/Jaspar
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TATGTCCATG-------
TAAATACATGTAAAATT |
|


|
|
MA0141.2_Esrrb/Jaspar
| Match Rank: | 9 |
| Score: | 0.62
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TATGTCCATG----
--TGACCTTGANNN |
|


|
|
NFAT(RHD)/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer
| Match Rank: | 10 |
| Score: | 0.61
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TATGTCCATG
ATTTTCCATT |
|

|
|